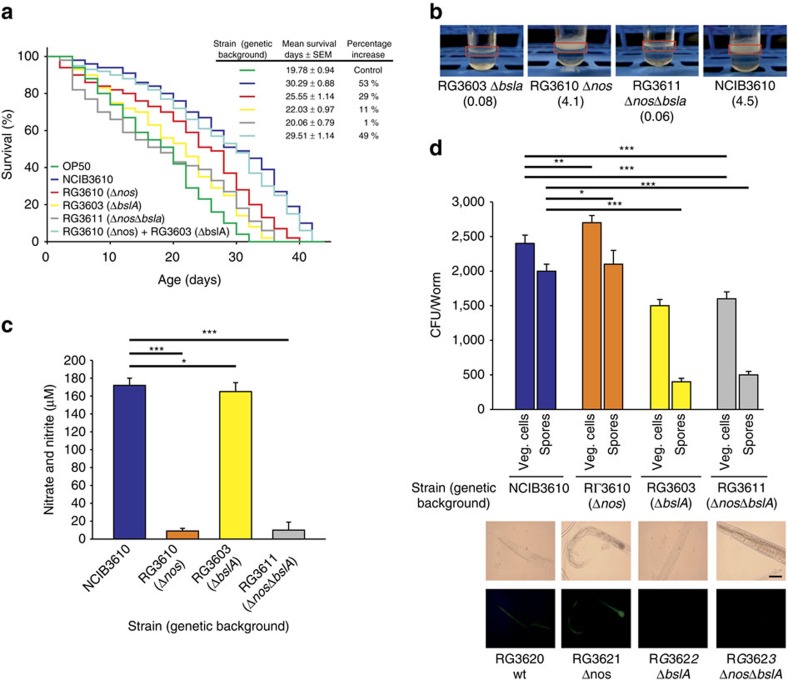
Figure 4. Biofilm formation and nitric oxide production are required to extend C. elegans lifespan.
(a) Effects upon longevity of worms fed on undomesticated B. subtilis defective in biofilm production and/or NO synthesis. Fifty L4 worms were fed either on spores of undomesticated NCIB3610 (blue) or RG3610 (Δnos, defective in NO production, orange) or RG3603 (ΔbslA, defective in biofilm synthesis, yellow) or RG3611 (ΔnosΔbslA, defective both in NO production and biofilm synthesis, grey) or a mixture of RG3610 and RG3603, light blue) B. subtilis on NGM agar plates, and survival was monitored. A representative experiment±s.e.m. is shown. (b) Biofilm formation proficiencies of undomesticated B. subtilis strains harbouring mutations in genes essential for biofilm formation and/or NO production. Biofilms were developed in liquid NGM. (c) NO quantification of undomesticated B. subtilis strains harbouring mutations in genes essential for biofilm formation and/or NO production. The nitrite and nitrate concentrations are expressed as μM. Error bars show the mean±s.e.m. (n=3). ***P<0.001 and *P<0.05 (analysis of variance (ANOVA) with Bonferroni test). (d) Colonization of the worm gut by B. subtilis strains deficient in biofilm formation and/or NO production. L4 worms were allowed to develop on NGM agar plates seeded with OP50 cells, washed and transferred to fresh NGM plates containing spores of the wild-type NCIB3610 (blue) or RG3610 (Δnos, defective in NO production, orange) or RG3603 (ΔbslA, defective in biofilm synthesis, yellow) or RG3611 (ΔnosΔbslA, defective both in NO production and biofilm synthesis, grey) B. subtilis strains, respectively. The worms were taken and processed to measure the number of B. subtilis cells in the worm gut, expressed as CFUs per worm. Error bars show the mean±s.e.m. (n=3). ***P<0.001, **P<0.01 and *P<0.05 (ANOVA with Bonferroni test). Images shown on the bottom are epifluorescence micrographs of typically stained N2 C. elegans fed epsA::gfp-expressing B. subtilis strains (RG3620, wild-type; RG3621, Δnos; RG3622, ΔbslA; and RG3623, ΔnosΔbslA). The fluorescence images were superimposed to differential interference contrast (DIC) images to depict the localization of the labels within the cells. Scale bar, 20 μm.