Figure 1.
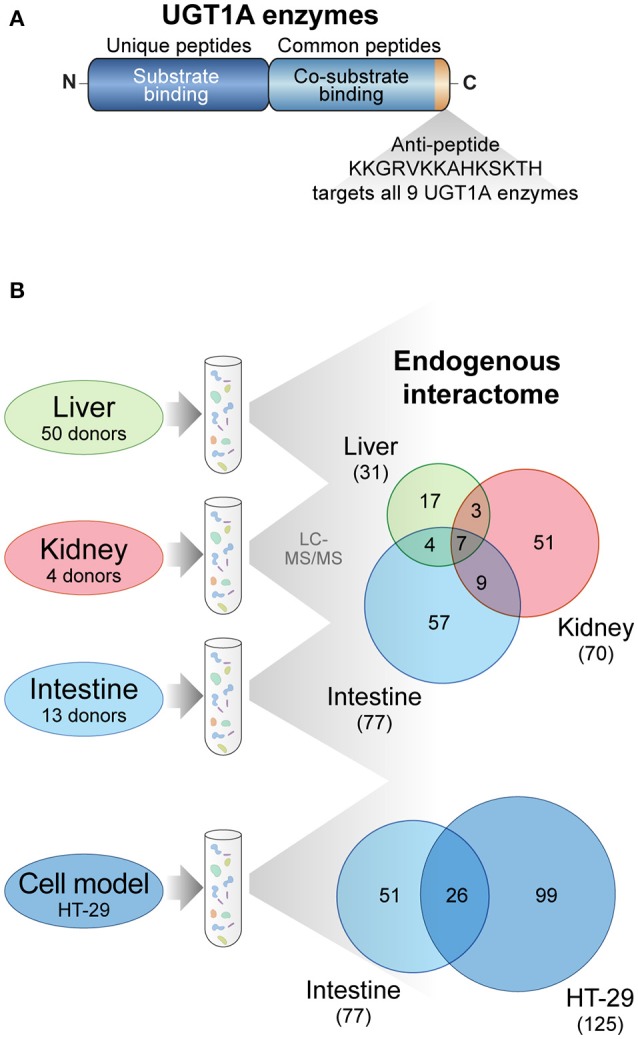
UGT1A interaction network investigated by untargeted proteomics. (A) The nine UGT1A enzymes are distinguished by the amino acid sequence of their substrate binding domain (unique peptides) whereas they share identical C-terminal co-substrate and transmembrane domains (common peptides). The anti-UGT1A antibody used in this study was raised against a C-terminal peptide common to all nine UGT1A enzymes but does not recognize the main spliced alternative isoforms 2 or UGT1A_i2s. (B) Experimental approach to establish endogenous UGT1A protein interactomes in drug metabolizing tissues and in the colon cancer cell model HT-29. Immunoprecipitation of UGT1A enzymes was conducted with the anti-UGT1A antibody. The numbers of common and unique UGT1A protein partners identified by mass spectrometry and above confidence threshold are represented in the Venn diagrams. Datasets were established on a minimum of two biological replicates. A Venn diagram for the 4 matrices is presented in Supplementary Figure 2. A list of proteins in each group is provided in Supplementary Table 3.
