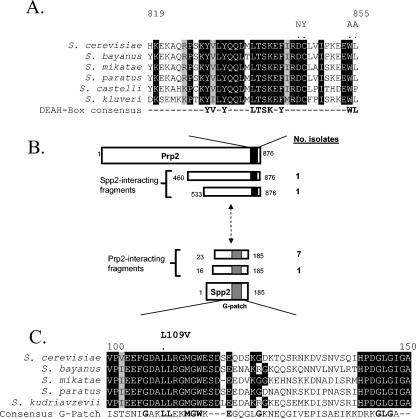
FIG. 1.
Spp2 interacts with the C terminus of Prp2. (A) Alignment of the region flanking the D845-W855 11-mer in Prp2 proteins among the Saccharomyces genus. The alignment was created by using the Pretty function in Seqweb 2.0 (Wisconsin GCG package) and was adjusted manually with sequences obtained from the www.yeastgenome.org website. Positions of complete conservation are indicated in black, and positions of strong similarity are outlined in gray. The positions of DC and WL are indicated with dots, and the DC/NY and WL/AA mutations are represented above the dots. A consensus derived from analysis of yeast DEAH-box splicing helicases is also shown. (B) Summary of yeast two-hybrid screening using full-length Prp2 or Spp2. The amino acid positions are indicated in the diagram, while the number of isolates of each prey fragment is listed at the right. The black box in Prp2 represents the D845-W855 11-mer and the flanking region. The gray box in Spp2 represents the G patch. (C) An alignment of the G patch of Spp2 of Saccharomyces spp. The amino acid positions in S. cerevisiae Spp2 are indicated. Positions of complete identity are in black, and positions with strong similarity are outlined in gray. The consensus G patch is shown as identified by the Conserved Protein Domain Database (26). Positions in boldface indicate agreement between Spp2 and the G-patch consensus.