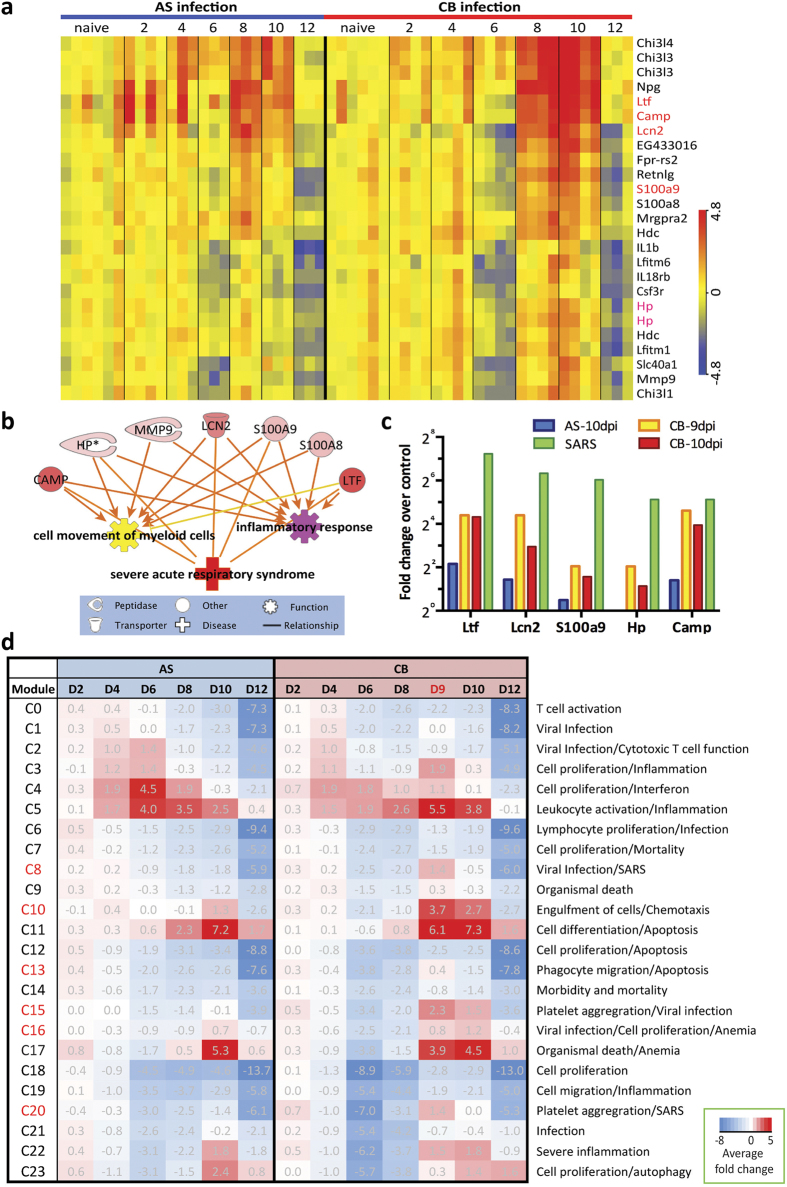
Figure 3. Hierarchical clustering and modular analyses reveal biological processes associated with severe malarial pathology.
(a) A set of probes representing 22 genes that were more highly up-regulated in CB than AS infection between 8–10 day post infection (dpi). They were identified from hierarchical clustering of all differentially expressed transcripts from any infected samples compared to naïve controls (related to Supplementary Fig. 1). Colour indicates the log 2 transformed normalized expression intensity. (b) Disease and Function analysis in IPA (Ingenuity Pathway Analysis) identified some of the 22 genes shown in (a) were significantly enriched in functions of ‘cell movement of myeloid cells’ and ‘inflammatory response’, and disease of ‘severe acute respiratory syndrome (SARS)’. (c) Five genes enriched in the SARS gene signature (red text in a) were even more highly up-regulated compared to naïve controls in the blood extracted from 4 CB infected mice that had reached humane end points at 9 dpi compared to randomly selected mice infected with AS or CB parasites at 10 dpi. Fold changes in SARS patients were from ref. 13. (d) The twenty-four modules identified by using self-organising map (SOM) method were presented with the average fold change of all differentially expressed transcripts within each module compared to naïve controls. Red represents positive mean fold change above zero (white) and blue indicates negative mean fold change. The clusters were annotated with IPA Diseases and Function analysis with manual curation. Clusters indicated in red showed greater up-regulation in CB infection during 9–10 dpi.