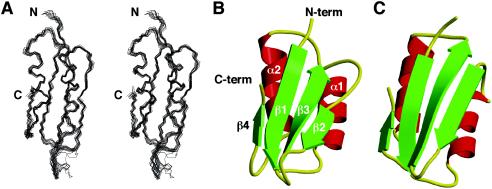
FIG. 1.
Structure of YbeD. (A) Stereo view of the backbone superposition of the 15 lowest-energy structures generated with MOLMOL (13). The superposition was done by using residues Phe11 to Leu87. (B) Ribbon representation of the YbeD structure generated with MOLSCRIPT (14) and Raster3D (19). The N terminus (N-term), C terminus (C-term), and secondary-structure elements are indicated. (C) The structure of the ACT domain of d-3-phosphoglycerate dehydrogenase (PDB code 1PSD) is very similar to that of YbeD, as backbone superposition of 48 residues resulted in 1.9-Å root-mean-square deviation.