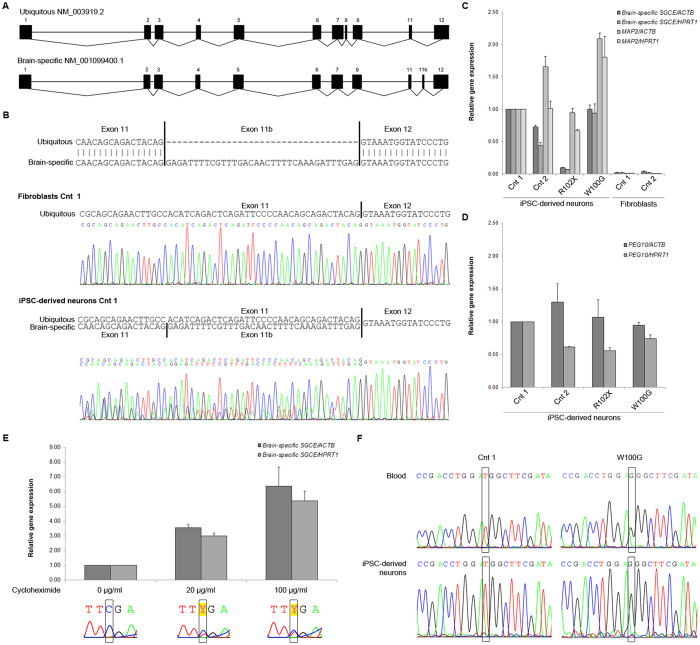
Figure 3. SGCE transcripts in fibroblasts and iPSC-derived neurons.
(A) Sequence alignment of the ubiquitous isoform (NM_003919.2) and the brain-specific isoform (NM_001099400.1). The two isoforms differ with respect to the presence of exons 8 and 11b. (B) cDNA sequencing indicates expression of the ubiquitous mRNA SGCE isoform in fibroblasts and the presence of the ubiquitous as well as the brain-specific isoform (includes exon 11b) in iPSC-derived neurons. (C) Gene expression analysis in iPSC-derived neurons from M-D patients and controls. The levels of the brain-specific SGCE transcript and the neuronal marker MAP2 were determined relative to the expression of the housekeeping genes ACTB and HPRT1. Fibroblasts from two healthy individuals were used as negative controls. Values were normalized to the neuronal control (Cnt) 1. The error bars indicate SE. (D) PEG10 expression analysis in iPSC-derived neurons from M-D patients and controls. The levels of PEG10 were determined relative to the expression of the housekeeping genes ACTB and HPRT1. The error bars indicate SE. (E) Expression of R102X brain-specific SGCE upon treatment with cycloheximide. Values were normalized to the nonsense mutant without treatment. The lower panel depicts the cDNA sequence upon different cycloheximide concentrations. (F) SGCE cDNA sequencing in blood and iPSC-derived neurons of the missense-mutant M-D patient (W100G). The absence of the maternal c.298T allele and selective expression of the paternal c.298G allele was in concordance with imprinting of the maternal wildtype allele in the patient neurons.