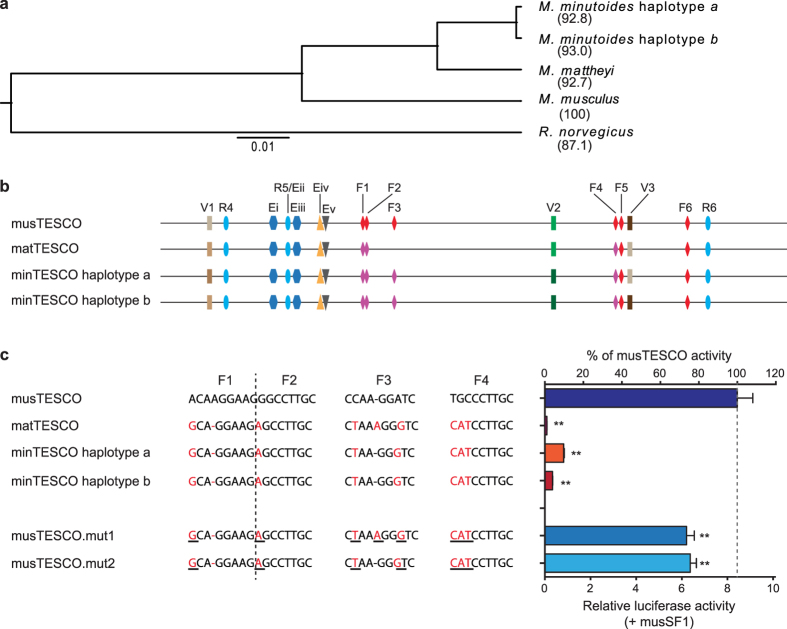
Figure 4. Markedly reduced enhancer activity of TESCO in M. mattheyi and M. minutoides.
(a) Maximum likelihood phylogeny using TESCO sequences. Numbers indicate percent sequence identity scores compared to musTESCO. (b) Schematic of TESCO enhancers from three Mus species. R4-6, SRY binding sites; F1-6, SF1 binding sites; Ei-v, evolutionarily conserved regions; V1-3, sequence variations between minTESCO a and b. Sequence changes at sites F1-4 may impair SF1 binding and are indicated with different colours. The F3 site predicted to have SF1 binding disrupted is removed from the matTESCO diagram. (c) Sequence comparison of F1-4 sites from the three species, and the mutants musTESCO.mut1/2. Sequence changes compared with musTESCO are in red and mutated sequences are underlined. Compared with musTESCO, mat/minTESCO showed markedly reduced activities in the presence of musSF1. Mutations of SF1 binding sites F1-4 in musTESCO to the corresponding sequence in matTESCO (musTESCO.mut1) or minTESCO (musTESCO.mut2) caused mildly reduced reporter activities in the presence of musSF1. Data are presented as TESCO luciferase activity normalized to co-transfected CMV-renilla luciferase activity. Error bars: s.e.m. (n = 3). Dashed lines indicate the levels of musTESCO activity. (**) P < 0.01 vs. musTESCO, one-way repeated measures ANOVA with Dunnett’s multiple comparisons test.