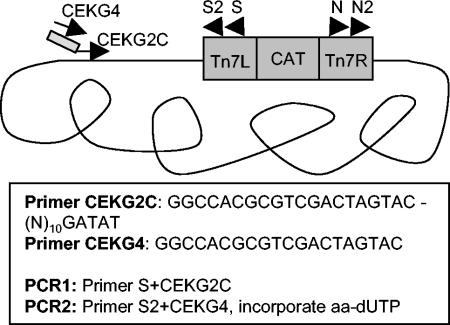
FIG. 1.
MATT method for labeling DNA adjacent to transposon insertions. The first PCR is run with genomic DNA template, primer S reading out from the left arm of the transposon and primer CEKG2C, which contains an anchored degenerate sequence. The second PCR uses the product of the first as template with nested transposon primer S2 and primer CEKG4, complementary to the conserved portion of primer CEKG2C. Amino-allyl dUTP is incorporated in the second PCR to allow conjugation of the resulting product to Cy3 or Cy5 fluorescent dyes for hybridization to the microarray.