Abstract
Adherence of bacteria to biotic or abiotic surfaces is a prerequisite for host colonization and represents an important step in microbial pathogenicity. This attachment is facilitated by bacterial adhesins at the cell surface. Because of their size and often elaborate multidomain architectures, these polypeptides represent challenging targets for detailed structural and functional characterization. The multifunctional fibrillar adhesin CshA, which mediates binding to both host molecules and other microorganisms, is an important determinant of colonization by Streptococcus gordonii, an oral commensal and opportunistic pathogen of animals and humans. CshA binds the high-molecular-weight glycoprotein fibronectin (Fn) via an N-terminal non-repetitive region, and this protein-protein interaction has been proposed to promote S. gordonii colonization at multiple sites within the host. However, the molecular details of how these two proteins interact have yet to be established. Here we present a structural description of the Fn binding N-terminal region of CshA, derived from a combination of X-ray crystallography, small angle X-ray scattering, and complementary biophysical methods. In vitro binding studies support a previously unreported two-state “catch-clamp” mechanism of Fn binding by CshA, in which the disordered N-terminal domain of CshA acts to “catch” Fn, via formation of a rapidly assembled but also readily dissociable pre-complex, enabling its neighboring ligand binding domain to tightly clamp the two polypeptides together. This study presents a new paradigm for target binding by a bacterial adhesin, the identification of which will inform future efforts toward the development of anti-adhesive agents that target S. gordonii and related streptococci.
Keywords: adhesin, bacterial pathogenesis, intrinsically disordered protein, microbiology, small-angle X-ray scattering (SAXS), X-ray crystallography
Introduction
The adherence of bacteria to material and host cell surfaces is essential for colonization, persistence, and pathogenicity. This process is facilitated by bacterial cell-surface components termed adhesins, which recognize and bind specific partner molecules presented on the surfaces of host cells and other microorganisms (1). The filamentous adhesins, comprising pili and fibrils, are among the largest and most complex of all bacterial surface proteins. They are found in both Gram-positive and Gram-negative bacteria, including many pathogenic species, and have attracted considerable attention due to their roles in colonization, infection, and as vaccine candidates (2, 3). Characteristically, these proteins form sizable polymeric assemblies that project outwards from the bacterial cell surface, presenting an adhesive target-binding region at their tip.
Fibrillar adhesins are expressed by a wide variety of bacteria and display an extraordinarily diverse array of adhesive functions. In contrast to the extensively studied pili, much still remains to be learned about the structures and functions of these proteins. Fibrillar adhesins usually include a single multidomain polypeptide chain, covalently tethered to the bacterial cell wall (1). They frequently display exquisite selectivity for their partner ligands, with their modular architectures providing a platform for multifunctionality (4–6). Although smaller than pili, they are inherently more complex, with divergent molecular structures, functions, and adhesive properties (7–9).
Streptococcus species produce a multitude of fibrillar adhesins that promote binding to host cells and other microorganisms. These include, among others, the M proteins of Streptococcus pyogenes and the antigen I/II family polypeptides (10–12). Streptococcus gordonii, a primary colonizer of the human oral cavity and an opportunistic pathogen, has been shown to present a number of fibrillar adhesins on its surface, including the polypeptide CshA. In vitro and in vivo binding assays, gene disruption experiments, and heterologous expression studies in the non-adherent bacterium Enterococcus faecalis have established CshA as an important determinant of S. gordonii adherence (4, 13). This protein has been shown to play a role in binding both to host molecules and a range of other microorganisms (14).
CshA is a 259-kDa polypeptide (4, 13) that forms ∼60-nm peritrichous fibrils on the surface of S. gordonii (4). It shares <10% overall sequence identity to any protein of known structure, indicative of divergent or novel function. The CshA pre-protein is composed of 2508-amino acid (aa)3 residues, organized in the form of a leader peptide (residues 1–41), a non-repetitive region (residues 42–778), 17 repeat domains (R1–R17, each ∼101-aa residues), and a C-terminal cell wall anchor (13). CshA has been shown to bind the high molecular weight glycoprotein fibronectin (Fn) via an interaction that is mediated by the N-terminal non-repetitive region of the protein (7). Antibodies specific to this portion of CshA block Fn binding, whereas those raised against the repeat domain region elicit no effect (15). The CshA-Fn interaction has been proposed to be of general significance in promoting S. gordonii colonization at a range of sites within the host (7, 15, 16). These include the cardiac endothelium, where adherence by S. gordonii is known to promote the onset of infective endocarditis, a severe, potentially fatal inflammation of the inner tissues of the heart (17).
Despite the importance of CshA, little is known about the molecular structure and function of this protein. Such information would provide fundamental mechanistic insight and may inform the development of anti-adhesive agents that target S. gordonii and related streptococci.
Here we report a structural and functional description of the non-repetitive Fn-binding region of CshA. We reveal that this part of the polypeptide is composed of three distinct domains, designated herein as non-repetitive domain 1 (NR1, CshA(42–222)), non-repetitive domain 2 (NR2, CshA(223–540)), and non-repetitive domain 3 (NR3, CshA(582–814)). In vitro Fn binding assays of truncated CshA proteins heterologously expressed on the surface of the non-adherent bacterium Lactococcus lactis demonstrate that both NR1 and NR2, but not NR3, confer adhesive properties to CshA. Biolayer interferometry analysis of Fn binding by the recombinant CshA NR domains reveals that NR2 binds both cellular and plasma fibronectin with significantly lower ka, kd, and KD values than NR1. Using circular dichroism (CD) spectroscopy, small angle X-ray scattering (SAXS), and allied biophysical methods, NR1 is shown to constitute a discrete intrinsically disordered domain (IDD). The crystal structure of NR2 is also presented, which adopts a lectin-like fold with a clearly identifiable ligand-binding site. Together, our data are consistent with a two-state mechanism of Fn binding by CshA, where NR1 functions to recognize and bind Fn, forming a dissociable pre-complex, which is subsequently stabilized by a high affinity binding interaction mediated by NR2. This “catch-clamp” mechanism of Fn binding may be of general significance in other bacterial adhesins that contain intrinsically disordered domains.
Results
Reassignment of CshA Domain Architecture
CshA has previously been shown to comprise four distinct regions as follows: an N-terminal signal peptide; a >80-kDa non-repetitive region; 17 ∼100-aa residue repeat domains, and a C-terminal cell wall anchor. In an effort to provide a more detailed description of the domain architecture of CshA, the sequence of this polypeptide was subjected to comprehensive bioinformatic analysis. Predictions of secondary and tertiary structure, disorder content, domain composition, and homology modeling of selected regions of the protein were performed. Our reassigned CshA domain composition is summarized in Fig. 1. The most significant consequence was assignment of the non-repetitive region of CshA into three distinct domains, designated NR1 (CshA(42–222)), NR2 (CshA(223–540)), and NR3 CshA(582–814)).
FIGURE 1.
Reassigned domain architecture of CshA. The CshA polypeptide comprises an N-terminal leader peptide (residues 1–40), a three-domain non-repetitive region (residues 41–819), a repeat region formed from 17 90–102-aa domains (residues 820–2507), and a C-terminal LPXTG cell wall anchor motif. The CshA domains NR1, NR2, NR3, and R13, produced recombinantly during this study, are highlighted (masses include vector encoded His6-tag and linker).
Fn Binding by CshA Is Confined to the NR1 and NR2 Domains
In previous studies, CshA has been shown to promote adherence by S. gordonii DL1 to the extracellular matrix protein Fn, an interaction mediated by the non-repetitive region of the protein (7, 14). To determine which of the NR domains of CshA contribute to Fn binding, full-length CshA (Fl_CshA) and two truncated forms of the protein, CshAΔNR1 and CshAΔNR1 + 2, were expressed on the surface of the non-adherent bacterium L. lactis. Genes encoding full-length CshA, CshAΔNR1, and CshAΔNR1 + 2 were cloned into the pMSP7517 shuttle vector (18) and transformed into wild type L. lactis by electroporation. Constructs encoding the two CshA deletion mutants included the wild type CshA leader peptide sequence fused to their N termini to ensure efficient export from the cytoplasm.
Levels of surface-expressed CshA polypeptides were determined by dot immunoblot analysis of S. gordonii DL1, L. lactis MG1363, and L. lactis strains expressing Fl_CshA, CshAΔNR1, or CshAΔNR1 + 2. Blots were probed with antibodies raised against the repetitive region of the protein (Fig. 2A). There was no reactivity of the CshA antiserum with the L. lactis MG1363 control. However, S. gordonii DL1 and each of the three L. lactis strains engineered to express CshA proteins presented the respective polypeptides on their surfaces. Variations in the levels of CshA surface expression were observed in each of the different strains, with Fl_CshA produced in a greater amount than CshAΔNR1 or CshAΔNR1 + 2 (Fig. 2A).
FIGURE 2.
CshA-dependent adherence of S. gordonii and L. lactis strains to immobilized cellular fibronectin. A, dot immunoblot analysis of CshA expression levels on the surface of S. gordonii and L. lactis strains. B, comparative levels of cFn adhesion by S. gordonii and L. lactis strains expressing CshA proteins. Mean values from four independent experiments are shown. Error bars represent standard deviations from the mean. *, p < 0.05; **, p < 0.006 as established by analysis of variance using Dunnett's multiple comparison test. ns, not significant.
To measure the relative binding properties of heterologously expressed Fl_CshA, CshAΔNR1, and CshAΔNR1 + 2, the ability of lactococci expressing each of these proteins to adhere to immobilized human cellular Fn (cFn) was assessed (Fig. 2B). Levels of adherence were compared with those of wild type S. gordonii DL1 and non-CshA-expressing L. lactis MG1363. Expression of full-length CshA on the surface of L. lactis conferred the ability to bind cFn. L. lactis cells expressing CshAΔNR1 were ∼2-fold more adherent to cFn than L. lactis::Fl_CshA, suggesting that exposure of NR2, by deletion of NR1, promotes target binding by CshA. L. lactis cells expressing CshAΔNR1 + 2 bound cFn at a level not significantly greater (as established by analysis of variance) than L. lactis MG1363, suggesting that the NR3 domain is dispensable for cFn binding.
Kinetics of Fn Binding by Recombinant CshA NR Domains
To further probe the Fn binding properties of CshA NR domains, each of NR1, NR2, and NR3 were recombinantly overexpressed in Escherichia coli and purified to homogeneity (>95% purity). Recombinant NR2 was isolated as a mixture of monomeric (∼70% of total material) and dimeric (∼30% of total material) species that could be readily separated by size-exclusion chromatography. Concentration of monomeric NR2 by ultrafiltration failed to yield the dimeric species, suggesting that NR2 dimerization is concentration-independent. Given this, and the absence of any prior experimental evidence to support homodimerization of the CshA non-repetitive region (4, 13), monomeric NR2 was taken to be the biologically relevant form of the protein and was used in all subsequent Fn binding studies.
The kinetics of human cFn and plasma Fn (pFn) binding by recombinant CshA NR domains were determined by biolayer interferometry (Blitz, ForteBio). Both cFn and pFn were analyzed as they share many structural features and functional properties but are synthesized by different cell types and at different locations. Fn proteins were amide-coupled to the tip surface of amine-reactive second-generation (AR2G) biosensors (ForteBio) prior to analysis. Binding affinities were measured between each of the two Fn proteins and NR1, monomeric NR2, and NR3; the control proteins BSA and CshA repeat domain 13 (R13); and a commercially sourced anti-Fn antibody (Dao; Fig. 3). The R13 domain of CshA was chosen for analysis as unlike other repetitive domains of the polypeptide, recombinant protein could be readily produced in high quantity. Experimentally determined kinetic parameters for cFn and pFn binding are summarized in Table 1. Neither BSA nor CshA R13 bound either form of Fn. By contrast, the anti-Fn antibody bound both substrates with an equilibrium dissociation constant (KD) in the low micromolar range (Table 1). Recombinant NR1 bound both cFn and pFn with a KD value approximately twice that of the anti-Fn antibody. This confirms that NR1 binds cFn and pFn but with lower affinity than the anti-Fn antibody. NR2 was found to bind cFn and pFn with a >10- and >4-fold lower KD value, respectively, than NR1, and a >5- and >2-fold lower KD value, respectively, than the anti-Fn antibody. These data demonstrate that NR2 binds Fn with high affinity, an observation in keeping with our heterologous expression studies, where deletion of NR1 serves to promote adherence to cFn by L. lactis strains. Kinetic parameters for Fn binding by CshA NR1 and NR2 suggest that NR1 forms a rapidly assembled (higher ka) but readily dissociable (higher kd) complex with Fn, whereas substrate binding by NR2 occurs over a longer time scale (lower ka), yielding a less rapidly dissociating complex (lower kd). Although there is variation in the kinetic parameters of cFn and pFn binding by individual CshA domains, the relative binding properties of each domain are consistent irrespective of the form of Fn used. NR3 is found to bind both cFn and pFn weakly, confirming a negligible role in defining the adhesive properties of CshA.
FIGURE 3.

Sensorgrams of fibronectin binding by CshA recombinant domains and control proteins. Representative of the binding of each CshA fragment (NR1, NR2, NR3, and R13), BSA and Fn antibody, to either plasma (A) or cellular (B) Fn. Note: the KD values are computationally calculated based on the exact amount of Fn deposited on the biosensor tip, which varies from run to run.
TABLE 1.
Kinetic parameters of cFn and pFn binding by recombinant CshA domains
No binding was observed between c/pFn and BSA.
| CshA domain | KD | ka | ka error | kd | kd error |
|---|---|---|---|---|---|
| μm | m−1s−1 | m−1s−1 | s−1 | s−1 | |
| cFn binding | |||||
| NR1 | 5.26 | 3.1 × 103 | 8.71 × 101 | 1.63 × 10−2 | 9.21 × 10−4 |
| NR2 | 0.5 | 1.38 × 103 | 1.35 × 101 | 6.89 × 10−4 | 1.05 × 10−5 |
| NR3 | 12.5 | 8.05 × 102 | 2.10 × 101 | 1.00 × 10−2 | 1.34 × 10−4 |
| Anti-Fn antibody | 1.94 | 6.29 × 103 | 2.95 × 102 | 1.22 × 10−2 | 3.61 × 10−4 |
| pFn binding | |||||
| NR1 | 3.45 | 9.07 × 103 | 6.67 × 102 | 3.13 × 10−3 | 9.11 × 10−4 |
| NR2 | 0.85 | 2.77 × 103 | 2.21 × 101 | 2.36 × 10−3 | 2.61 × 10−5 |
| NR3 | 9.42 | 1.33 × 103 | 3.73 × 101 | 1.27 × 10−2 | 2.31 × 10−4 |
| Anti-Fn antibody | 1.77 | 7.09 × 103 | 3.48 × 102 | 1.25 × 10−2 | 4.06 × 10−4 |
CshA NR1 Is a Discrete, Intrinsically Disordered Domain
Inspection of the aa sequence of the NR1 domain of CshA reveals that this protein is rich in disorder-promoting residues (10% Pro, 11% Glu, and 12% Ser) and deficient in order-promoting residues (<5% Ile and Leu, with no Cys, Phe, Trp, or Tyr). Analysis of the NR1 sequence using PONDR-FIT (19) suggests that >95% of NR1 is structurally disordered (Fig. 4A). To determine whether NR1 is indeed an IDD, the protein was subjected to biophysical and hydrodynamic characterization. Recombinant NR1 displays aberrant migratory behavior when analyzed by SDS-PAGE, characteristic of intrinsic disorder (20). The protein migrates with an apparent molecular mass of ∼37 kDa (Fig. 4B), although it possesses an experimentally verified mass of 20.8 kDa (including hexahistidine tag and linker; data not shown). Similarly, the CD spectrum of the protein is consistent with an absence of secondary structure, with a single minimum centered around 200 nm and no strong negative signals above 205 nm (Fig. 4C). Deconvolution of this spectrum employing the CDSSTR algorithm (21) and reference sets 6 and 7 (22, 23) gives a predicted disorder content of 79–81%.
FIGURE 4.
Biophysical and structural characterization of CshA NR1. A, PONDR-FIT analysis of the CshA NR1 aa sequence. B, SDS-PAGE analysis of recombinant NR1. The predicted molecular mass of the protein based on amino acid composition is 20.8 kDa. C, far-UV CD spectrum of CshA_NR1. The trace shown is a mean average calculated from 10 scans of the same protein sample. D, SAXS scattering profile of CshA NR1. A Kratky plot derived from the scattering profile is shown as an inset.
To further investigate the structure of NR1, this protein was analyzed by SAXS, a technique that is widely recognized as indispensable for the assignment and characterization of IDDs (24). SAXS data were processed to yield Guinier and Kratky plots and to calculate the shape and size of NR1 (Fig. 4D). The Guinier plot is linear in Q2 in the low angle region confirming that recombinant NR1 is monodisperse in solution (data not shown). Analysis of the Guinier region (RgQ <1.3) yields a radius of gyration of ∼67 Å, consistent with an extended conformation in solution. The Kratky plot derived from the NR1 scattering curve contains a distinct peak indicative of globular structure; however, the data do not progress to zero at high Q, suggestive of significant disorder content. Calculated dimensions of NR1 reveal an elongated structure of ∼250 × ∼70 Å in size, consistent with a loosely compacted but predominantly disordered structure. This contrasts with theoretically calculated dimensions of ∼560 × ∼2 Å for the fully disordered polypeptide.
Crystallization and Structure Determination of NR2
Having established that NR1 is an IDD, we next turned our attention to the NR2 domain of CshA. Both monomeric and dimeric forms of this protein were subjected to crystallization screening. Monomeric NR2 proved recalcitrant to crystallization; however, dimeric NR2 was readily crystallizable. The structure of this protein was determined to 2.7 Å resolution in space group P6222 using the single wavelength anomalous dispersion (SAD) method as applied to selenomethionine (SeMet)-labeled crystals of dimeric NR2. The asymmetric unit includes two copies of NR2, each of which forms a strand-swapped homodimer with a partner monomer from a neighboring asymmetric unit (Fig. 5A). Compelling electron density was not observed for the residues comprising the vector-encoded His6 tag and cleavable linker and the residues 313–331, 393–397, and 488–560 in either of the two copies of NR2. These residues have consequently been omitted from our final model. Inspection of the NR2 strand-swapped dimer reveals that each of the two monomers buries its three C-terminal β-strands (residues 447–488) into the central core of its dimer partner (Fig. 5A). Such extensive strand-swapping is unusual even in entirely β-strand proteins (25). In Fl_CshA, the N and C termini of NR2 are tethered to their neighboring NR1 and NR3 domains, respectively, an organization that would disfavor NR2 strand-swapping. Furthermore, previous studies of heterologously expressed Fl_CshA have revealed no evidence of dimer or higher oligomer formation (4, 13). Together, these observations imply that the observed dimerization is an artifact of protein overexpression. This provides further support for the validity of conducting our in vitro binding assays with monomeric recombinant NR2.
FIGURE 5.
Structural studies of CshA NR2. A, X-ray crystal structure of the CshA NR2 strand-swapped dimer. Individual monomers are colored yellow and purple, respectively. The location of loop regions containing residues 313–331 and 393–397 for which convincing electron destiny was not observed are indicated by dashed lines. B, structural model of CshA NR2 based on the strand-swapped dimer crystal structure. The location of the ligand-binding site and the associated capping loop are indicated. C, details of the CshA NR2 ligand-binding site, including electrostatic charge distribution (blue, positive; red, negative) and aa composition.
NR2 Is a Discrete Ligand Binding Domain with a Lectin-like Fold
Based on our NR2 strand-swapped dimer crystal structure, a model of the biologically relevant monomeric form of the protein was produced (Fig. 5, B and C). Inspection of this model revealed a globular protein with a lectin-like fold, comprising a β-sandwich core decorated with α-helices. The sandwich is formed from two anti-parallel β-sheets of four and five strands respectively, with the interface between the two sheets populated by predominantly hydrophobic amino acids. The β-sandwich is augmented by a series of short α-helices co-located at one end of the protein, which pack against strands 5, 7, and 10. The model suggests that the residues 313–331, for which compelling electron density was not observed in the NR2 dimer crystal structure, form a large flexible loop that caps a highly negatively charged pocket on the surface of the protein (Fig. 5, B and C). Given the absence of any other cavities, clefts, or highly charged surfaces, it is likely that this pocket constitutes the ligand-binding site of NR2.
Identification of Structural Relatives of NR2 Supports a Role in Carbohydrate Binding
Despite minimal sequence identity to any protein of known structure, a DALI search using monomeric NR2 identifies a number of structurally related proteins. These include the N1 region of the S. gordonii adhesin Sgo0707 (Z-score = 8.9 (26)), the adhesive V domains of the antigen I/II polypeptides SspB (Z-score = 7.1 (27)), and SpaP (Z-score = 7.0 (28)), and the endo-β-1,4-galactanase TmCBM61 from Thermotoga maritima (Z-score = 5.8 (29). Structural identity between NR2 and its relatives is predominantly restricted to the β-sandwich core of the protein, with significant divergence in the extent and identity of structural elements that decorate this sub-assembly (Fig. 6). Each of the structural homologues identified are involved in binding carbohydrates or glycoproteins, and with the exception of TmCBM61, they act as the adhesive component of a cell wall-anchored polypeptide in a Gram-positive bacterium.
FIGURE 6.
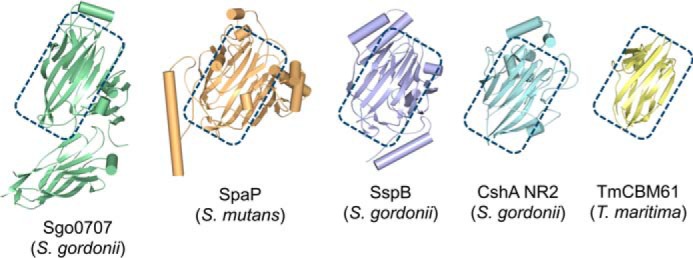
Comparison of CshA NR2 with structurally related homologues. Structures of the N-terminal domain of S. gordonii Sgo0707 (green), the AgI/II polypeptide V domains of Streptococcus mutans SpaP (orange) and S. gordonii SspB (blue), CshA_NR2 (model based on the strand-swapped dimer crystal structure; cyan), and T. maritima TmCBM61 (yellow) are shown. The conserved β-sandwich core of each protein is highlighted with a blue rectangle.
Discussion
In previous studies we have established the important role played by the S. gordonii cell wall-anchored polypeptide CshA in mediating adherence to host and microbial cell surfaces (4, 7, 14, 15). Here we extend the scope of our investigations to include detailed molecular level characterization of the Fn-binding “non-repetitive region” of this protein. This portion of CshA possesses an aa sequence distinct from that of any polypeptide for which structural or functional studies have been conducted to date, indicative of novel form or function.
In vitro cFn binding assays of heterologously expressed wild type CshA and truncated variants thereof confirm the critical role played by the non-repetitive region of CshA in facilitating cFn binding. These data also demonstrate that the adhesive properties of CshA are confined to the NR1 and NR2 domains of the protein. Surprisingly, a CshAΔNR1-expressing strain of L. lactis is more adherent to immobilized cFn than that expressing the wild type polypeptide. These data imply a mechanism of Fn binding by CshA where NR2 forms a dominant tight-binding interaction with its Fn substrate.
Results from complementary experiments investigating the kinetics of Fn binding by recombinant NR proteins are consistent with our heterologous expression studies. NR1 is found to bind both cFn and pFn with a KD value in the low micromolar range. NR2 binds both forms of Fn with greater affinity than NR1, demonstrating that this domain forms a stable, less dissociable interaction with Fn than its NR1 neighbor. Conversely, NR1 exhibits both a ka and kd value for c/pFn greater than that of NR2, indicating that NR1 engages and disengages Fn more rapidly. NR3, which was found to be dispensable for Fn binding in our heterologous expression studies, is shown using the BLitz technique to bind cFn and pFn only weakly, indicating that this domain may not significantly contribute to Fn binding in vivo. Together, our data are consistent with a mechanism of Fn binding by CshA that involves a rapidly formed but readily dissociable NR1-Fn complex, in tandem with a higher affinity, but less frequently formed NR2-dependent interaction.
To further probe the mechanism of Fn binding by CshA, recombinant NR1 and NR2 were subjected to structural characterization. NR1 is shown to exhibit all the hallmarks of an IDD, including aberrant hydrodynamic behavior and an absence of secondary structure (20, 30). SAXS analysis of NR1 demonstrates that the protein adopts a disordered but partially compacted structure in solution. Such a conformation is typical of IDDs, which form loosely assembled dynamic structures, stabilized by residual local and long-range weak intramolecular forces (31). This results in inherent conformational adaptability and, in the case of IDDs that are involved in protein or ligand binding, confers an expanded capture radius, enabling target binding over greater distances than can be achieved by globular proteins (32). Target engagement by IDDs may be concomitantly accompanied by partial or complete recovery of a fold state, a process that often brings both partners into closer proximity, potentially enhancing the rate of binding by the so-called “fly-casting” mechanism (33). Given that the energy needed to recover secondary structure is abstracted from the interaction energy of binding, IDDs generally form low affinity interactions with their binding partners, characterized by remarkably fast on- and off-rates (34). Fn binding by NR1 exhibits many of these defining characteristics, including rapid complex formation and dissociation and a weak yet specific relative binding affinity. Interestingly, NR1 is devoid of sequence motifs that have been previously implicated in IDD-mediated bacterial adherence to cell-surface molecules, implying that Fn binding by CshA NR1 may proceed via a mechanism that is distinct from, for example, the tandem β-zipper model (35). There was no evidence of NR1-NR2 complex formation, as assessed by native PAGE, when the two domains were mixed in vitro, implying that NR2 does not bind NR1 and act to template its folding.
In contrast to NR1, the NR2 domain of CshA is shown to adopt a globular structure with a lectin-like fold. The protein presents a clearly identifiable ligand-binding site on its surface that is populated by a combination of aromatic and negatively charged residues. This composition is consistent with a role for NR2 in the binding of carbohydrates or glycoproteins, a function further supported by our in vitro binding data, and the identification of structural homologues of NR2 that mediate protein-carbohydrate interactions (26–29, 36). The ligand-binding site of NR2 is capped by a sizeable loop, for which no electron density was observed during structure elucidation. The absence of compelling electron density in this region is indicative of flexibility. It therefore appears likely that the capping loop plays a role in gating access to the NR2-binding site, potentially acting to expose this highly charged pocket during Fn engagement. Based on available structural and functional data, it is not possible to unambiguously identify the specific glycosylation(s) upon Fn that are bound by NR2, although this represents a major focus for future investigations.
In summary, we report herein a structural and functional dissection of the Fn-binding, non-repetitive region of the S. gordonii fibrillar adhesin CshA. Uniquely, CshA appears to employ a bipartite mechanism of target binding that involves the coordinated action of a low affinity but high capture radius IDD, in partnership with a high affinity, high specificity ligand binding domain. Our data suggest a two-step catch-clamp Fn-binding mechanism, wherein the disordered NR1 domain of CshA acts to engage and “catch” Fn, via a process that is expedited by its elongated, disordered conformation. The resulting NR1-Fn pre-complex is rapidly formed and dissociated, although it may be stabilized via an NR2-mediated tight binding interaction, which functions to “clamp” CshA and Fn together (Fig. 7). Such a mechanism offers an elegant solution to the problem of forming highly specific intermolecular interactions within molecularly rich environments such as the human host. A number of fibrillar adhesins have been identified that incorporate IDDs, and as such the catch-clamp mechanism outlined herein may be of general applicability to other bacterial surface proteins that possess such domains (37). Finally, the unusual target binding mechanism exhibited by CshA, along with molecular insights provided herein, presents a framework for the development of new anti-adhesive interventions that target disease causing streptococci and related bacteria. Such interventions could be based on multicomponent agents that target both the IDD and ligand-binding steps during adherence.
FIGURE 7.
Schematic depicting the catch-clamp mechanism of Fn binding by CshA. The intrinsically disordered NR1 domain of the protein rapidly engages and binds Fn in a process expedited by the sizable capture radius of the domain. Fn binding results in the formation of a dissociable pre-complex that may be accompanied by the recovery of NR1 secondary structure. The resulting pre-complex is stabilized by a high affinity binding interaction mediated by the NR2 domain of CshA.
Experimental Procedures
Bioinformatic Analysis and Reassignment of CshA Domain Architecture
Bioinformatic analysis and homology modeling studies were performed using non-proprietary software or publicly accessible web servers. In cases where the CshA amino acid sequence was too long for analysis, the protein sequence was sub-divided into appropriately sized regions and each was analyzed independently. Domain composition of CshA and the identification of domain boundaries were performed using Interpro (38) integrating 11 databases, including Pfam (39), PRINTS (40), PROSITE (41), and ProDom (42), to provide prediction of protein families and domains. Complementary analyses were performed using the DomPred server (43). Disorder prediction was performed using PONDR-FIT (19), secondary structure prediction performed using PSIPRED (44), and signal peptide prediction performed using RPSP (45), and the presence of coiled-coils was established using COILS (46). Where a suitable template or templates could be identifies, SWISS-MODEL (47–50) or PHYRE2 (51) were used to generate homology models of identified CshA domains to allow more accurate assignment of domain boundaries.
Bacterial Strains and Growth Conditions
S. gordonii DL1 (Challis) (52) was routinely cultured in BHY broth (3.7% brain-heart infusion (LabM), 0.5% yeast extract (Bacto)), anaerobically at 37 °C. L. lactis MG1363 was cultured in GM17 broth (3.72% M17 (Difco), 0.5% glucose) at 30 °C under anaerobic conditions. Cultures of L. lactis expressing CshA, CshAΔNR1, or CshAΔNR1 + 2 were supplemented with 5 μg/ml erythromycin. E. coli Gigasingles (EMD Millipore) and BL21 were cultured in LB broth (2.5% Luria-Bertani (Fisher Scientific)) at 37 °C with shaking at 200 rpm, supplemented with 50 μg/ml carbenicillin.
Generation of L. lactis Heterologous Expression Strains
Plasmids (Tables 2 and 3) or PCR amplicons were purified using QIAquick Spin Miniprep or PCR purification kits, respectively (Qiagen). Oligonucleotides were synthesized by MWG Eurofins. Chromosomal DNA was extracted from S. gordonii DL1 as described previously (53). Unless otherwise stated, DNA was PCR-amplified with PrimeSTAR GXL DNA polymerase (Clontech). Primers pMSP-cshA.F/R (Tables 2 and 3) were designed to amplify the entire cshA gene from S. gordonii DL1 chromosomal DNA, incorporating unique NcoI/XhoI restriction endonuclease sites at its termini. The purified amplimer was cloned into vector pMSP7517 (18) using ligation-independent In-Fusion HD cloning kit (Clontech). The resultant construct (pMSP::cshA) was transformed into competent E. coli StellarTM cells (Clontech), recovered, and confirmed by sequencing. This construct was employed as template for in-frame deletion of NR1 (primers cshA.Rev2/Fwd2) or NR1 + 2 (primers cshARev.primerA/cshAFwd.primerB) from cshA, based upon a circular PCR method (54) with 5′-phosphorylated primers. Following PCR amplification with Phusion High-Fidelity DNA polymerase (New England Biolabs), template was removed by DpnI digestion, and purified amplimers were self-ligated using T4 DNA ligase. Constructs were transformed into E. coliStellarTM cells and confirmed by sequencing. Consequently, the three pMSP-derived constructs were electroporated into L. lactis MG1363, as described previously (55). Transformants were recovered on GM17 agar supplemented with erythromycin, and expression of CshA, CshAΔNR1, or CshAΔNR1 + 2 on the surface of L. lactis was verified by SDS-PAGE and Western immunoblot analyses.
TABLE 2.
Primers used during this study
fwd means forward, and rev means reverse.
| Primer name | Sequence 5′–3′ | Nucleotide no. |
|---|---|---|
| CshA-Head1_fwd | aagttctgtttcagggcccgatgGATGAAACAAGCGCTTCTGGTGTTCAA | 124–150 |
| CshA-Head1_rev | atggtctagaaagctttactactaGTTAATTTGTTTGGCTAATTGAGTACT | 639–666 |
| CshA-Head2_fwd | aagttctgtttcagggcccgatgTGGGTTGATTTTTCTGATACAGCAAGT | 666–693 |
| CshA-Head2_rev | atggtctagaaagctttactactaCTCACGGTCATCGGCCGTCCAGTCACT | 1594–1620 |
| CshA-Neck1_fwd | aagttctgtttcagggcccgatgAATGGTAATGCCAAGGCCTATGTCAAA | 1621–1770 |
| CshA-Neck2_rev | atggtctagaaagctttactactaACCAGTCGAATCGTATTTTGGTACCTT | 2416–2442 |
| CshA-R13_fwd | aagttctgtttcagggcccgatgACAGGAACTGGTGCAACTAGCACAGGT | 6094–6120 |
| CshA-R13_rev | atggtctagaaagctttactactaTACTTTCGTAAACTCTGGACTGTAGGT | 6364–6390 |
| pMSP-cshA.F | GAGGCACTCACCATGGAGGAGGTCATTATGGGAAAAGATTTG | −5–15a |
| pMSP-cshA.R | GGAGACCGGCCTCGAGATACAGGACAGAAAACCCTTC | +1254–+1274a |
| cshA.Rev2 | P-TTCATCAGCGCTAACCTG | 112–129 |
| cshA.Fwd2 | P-TGGGTTGATTTTTCTGATACAG | 667–688 |
| cshARev.primerA | P-TTCATCAGCGCTAACCTGCGTTGC | 106–129 |
| cshAFwd.primerB | P-CGTGAGGATGTAGCCGATGAAGGT | 1615–1638 |
a − are base pairs upstream of cshA gene; + are base pairs downstream of the cshA gene.
TABLE 3.
Plasmids used during this study
Western Immunoblotting Analysis
Expression of CshA, CshAΔNR1, or CshAΔNR1 + 2 on the surface of L. lactis was induced by culturing cells in the presence of 10 ng of nisin/ml. Suspensions were adjusted to A600 nm 1.5 and spotted (2 μl) onto nitrocellulose membrane. The membrane was blocked with TBS (50 mm Tris-HCl, pH 7.6, 0.15 m NaCl) containing 10% (w/v) milk powder, and subsequently probed with rabbit α-CshA-RR (15) followed by swine anti-rabbit IgG-horseradish peroxidase (HRP) conjugate (Dako), both diluted 1 in 1000 into TBS supplemented with 0.1% Tween 20 and 1% milk powder. The membrane was then developed using Amersham BiosciencesTM ECLTM Western blotting analysis system (GE Healthcare) according to manufacturer's instructions.
Cell Binding Assays
Human cFn (Sigma) was diluted in coating buffer (20 mm Na2CO3, adjusted to pH 9.3 with NaHCO3) and adsorbed onto microtiter plate wells (Immulon 2HB) for 16 h at 4 °C. Nonspecific binding sites were blocked with 3% BSA containing 0.05% Tween, and the wells were washed with TBSC (10 mm Tris-HCl, pH 7.6, 150 mm NaCl, 5 mm CaCl2). Bacterial cells in TBSC (5 × 108 cells/ml) were added to wells (0.1 ml) and incubated for 2 h at 37 °C. Adhered cells were fixed, stained with crystal violet, and biomass quantified by measuring A595 (8).
Gene Cloning
DNA fragments corresponding to CshA(42–222) (NR1), CshA(223–540) (NR2), CshA(582–814) (NR3), and CshA(1032–2130) (R13) were PCR-amplified from either plasmid pAM401::cshA (4) or S. gordonii DL1 chromosomal DNA using primers listed in Tables 2 and 3. PCR products were ligated into vector pOPINF (56) using the In-FusionTM system (Clontech). Resulting plasmids encoded N-terminally hexa-histidine-tagged CshA domains. All constructs was verified by DNA sequencing.
Protein Expression and Purification
E. coli BL21 (DE3) cells harboring CshA domain expression plasmids were grown in LB medium supplemented with carbenicillin at 37 °C to A600 = 0.4–0.6. Protein expression was induced by adding IPTG (1 mm) and incubating cultures for 16 h at 18 °C. Bacteria were harvested by centrifugation, suspended in 20 mm Tris-HCl, 150 mm NaCl, pH 8, and lysed with a cell disruptor (Z Plus Series cell disruptor, Constant Systems Ltd.) at 25,000 p.s.i. Cell supernatants were applied to a 5-ml Hi-Trap chelating column (pre-loaded with nickel, GE Healthcare) and eluted with an imidazole gradient (10–500 mm) over 15 column volumes. Fractions containing CshA proteins were pooled, concentrated, and further purified by passage through a Hi-Load 16/60 Superdex 75 column (GE Healthcare) pre-equilibrated in 20 mm Tris-HCl, 150 mm NaCl, pH 7.5. Fractions containing CshA proteins were pooled and concentrated by ultrafiltration to 10 mg/ml.
For the production of selenomethionine-labeled NR2, a culture of E. coli BL21 (DE3) cells harboring pOPINF:cshA-NR2 was grown in LB medium supplemented with carbenicillin at 37 °C for 16 h. Cells were harvested by centrifugation, washed three times, suspended in double-distilled water, and used to inoculate 1 liter of SelenoMetTM medium (Molecular Dimensions Ltd.) containing carbenicillin, supplemented with SelenoMet Nutrient Mix (Molecular Dimensions Ltd.). The culture was grown at 37 °C with shaking to A600 = 0.6. A mix of amino acids was added to the culture (100 mg of lysine, 100 mg of phenylalanine, 100 mg of threonine, 50 mg of isoleucine, 50 mg of leucine, 50 mg of valine, 60 mg of selenomethionine), which was then incubated with shaking, for 15 min, at 37 °C. IPTG was added (final concentration 1 mm) and the culture incubated for 16 h at 18 °C. Cells were harvested and processed for protein purification as above with the addition of 2 mm tris(2-carboxyethyl)phosphine to all chromatography buffers.
Mass Spectrometry
For accurate intact mass determination, a sample of recombinant CshA NR1 with the His tag intact was prepared. Mass analysis was performed using an UltrafleXtreme mass spectrometer (Bruker Daltonik) in positive reflectron ion mode. Data analysis was performed using the Bruker Flex Analysis software.
Biolayer Interferometry
The BLItz® system (ForteBio) was used to measure binding affinity and kinetics of the interaction between cFn or pFn with CshA recombinant protein fragments. All experiments were performed at 20 °C, using PBS-based, pH 7.4, Kinetics Buffer (ForteBio). c/pFn were immobilized onto Dip and ReadTM Amine Reactive Second-Generation (AR2G) biosensors using N-hydroxysuccinimide/1-ethyl-3-(3dimethylaminopropyl)carbodiimide (NHS/EDC)-catalyzed amide bond formation at 20 μg/ml in 10 mm acetate buffer, pH 4.0, for 5 min. Probes were quenched in 1 m ethanolamine, pH 8.5, and used for kinetic assays of CshA protein activities. The association and dissociation kinetics of the CshA proteins were performed at concentrations of 5, 15, 25, and 50 μm. The kinetic dataset was globally fitted employing a 1:1 binding model.
CD Spectroscopy
CD spectra of 1 mg/ml recombinant NR1 in 20 mm phosphate, pH 7.5, 150 mm NaCl, were collected using an Aviv model 410 CD spectrometer at 20 °C, using a 0.05-cm path length cuvette, with a wavelength interval of 1 nm and a data collection time of 1 s. The buffer-only spectrum was subtracted, and the data were converted to mean residue ellipticity. The CDSSTR algorithm (22, 23) and associated reference data sets were used for data analysis, accessed via the DichroWeb server (21).
Small Angle X-ray Scattering
SAXS data of CshA NR1 were collected at Diamond Light Source (beamline I22) at a wavelength of λ = 1 Å and a sample-to-detector distance of 2.5 m. Three sample concentrations were measured: 8, 4, and 2 mg/ml. Samples were exposed for 200 frames of 1 s. Each frame was sector-integrated with in-house beamline software. There was no evidence of concentration-dependent effects; thus data collected for the 8 mg/ml sample of NR1 were used for further analysis. Buffer scattering was subtracted using in-house software Scientific Data Analysis for non-crystalline diffraction. Particle distance distribution function (p(r)) and maximum intra-particle dimension (Dmax) were calculated with GNOM (57). PRIMUS (58) was used to calculate the radius of gyration (Rg) employing the Guinier approximation and to generate a Kratky plot. The shape of the protein was evaluated using DAMMIF (59).
Protein Crystallization
Conditions supporting the growth of crystals of native and SeMet-labeled dimeric CshA NR2 were initially identified using the hanging drop method of vapor diffusion at 20 °C and commercially available screens. Diffraction quality crystals of unlabeled or SeMet-labeled CshA NR2 were grown in 0.2 m Na/K tartrate, 20–25% polyethylene glycol 3350, MMT buffer (dl-malic acid, MES, and Tris base in the molar ratios 1:2:2), pH 5.0. Crystals selected for diffraction data collection were mounted in appropriately sized litholoops (Molecular Dimensions Ltd) and flash-cooled in liquid nitrogen without additional cryoprotection prior to analysis.
Diffraction Data Collection and Structure Determination
Diffraction data were collected at Diamond Light Source, UK, on beamlines I03 and I04-1. Data were processed with iMosflm (60) and scaled with Scala (61) as implemented in the CCP4 suite (62). Two sets of CshA_NR2 SeMet-labeled data were integrated using iMosflm and scaled with Scala. These data were then prepared for merging and scaling in Pointless (61, 63) and merged with Aimless (61, 64) to scale together multiple observations of reflections. The structure of CshA_NR2 was initially determined in space group P6222, with two molecules in the asymmetric unit, to 3.5 Å resolution, using the SAD method as applied to SeMet-labeled crystals of dimeric NR2. Identification of heavy atom sites and the resulting initial phase calculation was carried out using Phenix Autosol (65). 18 selenium sites were located with a figure of merit of 0.35. The output model was refined utilizing Refmac5 and employed as a molecular replacement search model to elucidate a higher resolution NR2 structure (2.7 Å) using PhaserMR (66), employing diffraction data collected from a crystal of unlabeled NR2. The resulting model was subjected to additional cycles of automated model building with Phenix AutoBuild (67), followed by iterative rounds of manual model building and refinement using COOT (68) and Refmac5 (69), respectively. Data collection, phasing, and refinement statistics for CshA NR2 are provided in Table 4. Protein structure figures have been prepared with PyMOL (Schrödinger, LLC).
TABLE 4.
Summary of X-ray data collection and refinement statistics
| CshA_NR2 | CshA_NR2-SeMeta | |
|---|---|---|
| Data collection | ||
| Space group | P6222 | P6222 |
| Cell dimensions | ||
| a, b, c (Å) | 173.93, 173.93, 104.07 | 172.60, 172.60, 101.85 |
| Resolution (Å) | 61.01–2.66 (2.8–2.66)a | 60.13–3.21 (3.43–3.21) |
| Rmerge | 10.7 (147.7) | 26.7 (129.1) |
| I/σI | 19.8 (2.1) | 16.3 (1.1) |
| CC1/2 (%) | 90.5 (39.1) | |
| Completeness (%) | 100.0 (100.0) | 96.5 (80.7) |
| Redundancy | 16.8 (16.6) | 36.0 (6.6) |
| No. of selenium sites | 18 | |
| Figure of merit | 0.35 | |
| Refinement | ||
| Resolution (Å) | 61.01–2.66 | |
| No. of reflections | 25,890 | |
| Rwork/Rfree | 19.5/25.2 | |
| No. of atoms | ||
| Protein | 7468 | |
| Ligand/ion | 0 | |
| Water | 2 | |
| B factors Å2 | ||
| Protein | 48.0 | |
| Ligand/ion | ||
| Water | 52.3 | |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.016 | |
| Bond angles (°) | 1.88 | |
| Ramachandran favored (%) | 92.8 | |
| Ramachandran outliers (%) | 2.3 | |
a Two separate CshA_NR2-SeMet crystals were used to collect data.
b Values in parentheses are for highest resolution shell.
Electrostatic Potential Calculations
For figures showing an electrostatic potential projected on the molecular surface of protein structures, the Poisson-Boltzmann electrostatic potential on the solvent-accessible surface is shown, with potential values ranging from −15 kT/e (red) to 15 kT/e (blue). This was calculated using the APBS plug-in (70) in PyMOL and a PQR file generated using PDB2PQR (71), with PARSE force-field charges after protonation state assignment by PropKa (72, 73).
Author Contributions
R. J. L., H. F. J., A. H. N., and P. R. R. conceived and designed the study. C. R. B., M. N. S., M. T., A. H. N., and P. R. R. performed experiments and analyzed data. C. R. B., M. N. S., R. J. L., H. F. J., A. H. N., and P. R. R. wrote the manuscript. R. J. L., H. F. J., and P. R. R. acquired funding. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Jane Brittan and Lindsay Dutton for technical assistance, Dr. Christopher Arthur for mass spectrometry analysis, and Kristian Le Vay, Genevieve Baker, and Dr Steven Burston for fruitful discussions.
This work was supported in part by National Institutes of Health Grants DE016690 (to H. F. J. and R. J. L.) and DE012505 (to R. J. L.), Biotechnology and Biological Sciences Research Council Grant BB/I006478/1 (to P. R. R.), and Royal Society University Research Fellowship Award UF080534 (to P. R. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors′ Picks.
The atomic coordinates and structure factors (code 5L2D) have been deposited in the Protein Data Bank (http://www.pdb.org/).
- aa
- amino acid
- R13
- repeat region 13
- IDD
- intrinsically disordered domain
- SeMet
- selenomethionine
- SAD
- single anomalous dispersion
- SAXS
- small angle X-ray scattering
- c/pFn
- cellular/plasma fibronectin
- NR
- non-repetitive
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside.
References
- 1.Kline K. A., Fälker S., Dahlberg S., Normark S., and Henriques-Normark B. (2009) Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5, 580–592 [DOI] [PubMed] [Google Scholar]
- 2.Demuyser L., Jabra-Rizk M. A., and Van Dijck P. (2014) Microbial cell surface proteins and secreted metabolites involved in multispecies biofilms. Pathog. Dis. 70, 219–230 [DOI] [PubMed] [Google Scholar]
- 3.Löfling J., Vimberg V., Battig P., and Henriques-Normark B. (2011) Cellular interactions by LPxTG-anchored pneumococcal adhesins and their streptococcal homologues. Cell. Microbiol. 13, 186–197 [DOI] [PubMed] [Google Scholar]
- 4.McNab R., Forbes H., Handley P. S., Loach D. M., Tannock G. W., and Jenkinson H. F. (1999) Cell wall-anchored CshA polypeptide (259 kilodaltons) in Streptococcus gordonii forms surface fibrils that confer hydrophobic and adhesive properties. J. Bacteriol. 181, 3087–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larson M. R., Rajashankar K. R., Patel M. H., Robinette R. A., Crowley P. J., Michalek S., Brady L. J., and Deivanayagam C. (2010) Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of α- and PPII-helices. Proc. Natl. Acad. Sci. U.S.A. 107, 5983–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruszka D. T., Wojdyla J. A., Bingham R. J., Turkenburg J. P., Manfield I. W., Steward A., Leech A. P., Geoghegan J. A., Foster T. J., Clarke J., and Potts J. R. (2012) Staphylococcal biofilm-forming protein has a contiguous rod-like structure. Proc. Natl. Acad. Sci. U.S.A. 109, E1011–E1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakubovics N. S., Brittan J. L., Dutton L. C., and Jenkinson H. F. (2009) Multiple adhesin proteins on the cell surface of Streptococcus gordonii are involved in adhesion to human fibronectin. Microbiology 155, 3572–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakubovics N. S., Kerrigan S. W., Nobbs A. H., Strömberg N., van Dolleweerd C. J., Cox D. M., Kelly C. G., and Jenkinson H. F. (2005) Functions of cell surface-anchored antigen I/II family and Hsa polypeptides in interactions of Streptococcus gordonii with host receptors. Infect. Immun. 73, 6629–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corrigan R. M., Rigby D., Handley P., and Foster T. J. (2007) The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153, 2435–2446 [DOI] [PubMed] [Google Scholar]
- 10.Fischetti V. A. (1989) Streptococcal M-protein–molecular design and biological behavior. Clin. Microbiol. Rev. 2, 285–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenkinson H. F., and Demuth D. R. (1997) Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol. Microbiol. 23, 183–190 [DOI] [PubMed] [Google Scholar]
- 12.Rego S., Heal T. J., Pidwill G. R., Till M., Robson A., Lamont R. J., Sessions R. B., Jenkinson H. F., Race P. R., and Nobbs A. H. (2016) Structural and functional analysis of cell wall-anchored polypeptide adhesin BspA in Streptococcus agalactiae. J. Biol. Chem. 291, 15985–16000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNab R., Jenkinson H. F., Loach D. M., and Tannock G. W. (1994) Cell-surface-associated polypeptides CshA and CshB of high-molecular-mass are colonization determinants in the oral bacterium Streptococcus gordonii. Mol. Microbiol. 14, 743–754 [DOI] [PubMed] [Google Scholar]
- 14.Holmes A. R., McNab R., and Jenkinson H. F. (1996) Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect. Immun. 64, 4680–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNab R., Holmes A. R., Clarke J. M., Tannock G. W., and Jenkinson H. F. (1996) Cell surface polypeptide CshA mediates binding of Streptococcus gordonii to other oral bacteria and to immobilized fibronectin. Infect. Immun. 64, 4204–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giomarelli B., Visai L., Hijazi K., Rindi S., Ponzio M., Iannelli F., Speziale P., and Pozzi G. (2006) Binding of Streptococcus gordonii to extracellular matrix proteins. FEMS Microbiol. Lett. 265, 172–177 [DOI] [PubMed] [Google Scholar]
- 17.Douglas C. W., Heath J., Hampton K. K., and Preston F. E. (1993) Identity of viridans streptococci isolated from cases of infective endocarditis. J. Med. Microbiol. 39, 179–182 [DOI] [PubMed] [Google Scholar]
- 18.Hirt H., Erlandsen S. L., and Dunny G. M. (2000) Heterologous inducible expression of Enterococcus faecalis pCF10 aggregation substance Asc10 in Lactococcus lactis and Streptococcus gordonii contributes to cell hydrophobicity and adhesion to fibrin. J. Bacteriol. 182, 2299–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue B., Dunbrack R. L., Williams R. W., Dunker A. K., and Uversky V. N. (2010) PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 1804, 996–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habchi J., Tompa P., Longhi S., and Uversky V. N. (2014) Introducing protein intrinsic disorder. Chem. Rev. 114, 6561–6588 [DOI] [PubMed] [Google Scholar]
- 21.Whitmore L., and Wallace B. A. (2008) Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 22.Compton L. A., and Johnson W. C. (1986) Analysis of protein circular-dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 155, 155–167 [DOI] [PubMed] [Google Scholar]
- 23.Manavalan P., and Johnson W. C. (1987) Variable selection method improves the prediction of protein secondary structure from circular-dichroism spectra. Anal. Biochem. 167, 76–85 [DOI] [PubMed] [Google Scholar]
- 24.Bernadó P., Mylonas E., Petoukhov M. V., Blackledge M., and Svergun D. I. (2007) Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 129, 5656–5664 [DOI] [PubMed] [Google Scholar]
- 25.Gronenborn A. M. (2009) Protein acrobatics in pairs–dimerization via domain swapping. Curr. Opin. Struct. Biol. 19, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nylander Å., Svensäter G., Senadheera D. B., Cvitkovitch D. G., Davies J. R., and Persson K. (2013) Structural and functional analysis of the N-terminal domain of the Streptococcus gordonii adhesin Sgo0707. PLoS ONE 8, e63768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forsgren N., Lamont R. J., and Persson K. (2009) Crystal structure of the variable domain of the Streptococcus gordonii surface protein SspB. Protein Sci. 18, 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Troffer-Charlier N., Ogier J., Moras D., and Cavarelli J. (2002) Crystal structure of the V-region of Streptococcus mutans antigen I/II at 2.4 angstrom resolution suggests a sugar preformed binding site. J. Mol. Biol. 318, 179–188 [DOI] [PubMed] [Google Scholar]
- 29.Cid M., Pedersen H. L., Kaneko S., Coutinho P. M., Henrissat B., Willats W. G., and Boraston A. B. (2010) Recognition of the helical structure of β-1,4-galactan by a new family of carbohydrate-binding modules. J. Biol. Chem. 285, 35999–36009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uversky V. N. (2013) A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 22, 693–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uversky V. N. (2015) Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 282, 1182–1189 [DOI] [PubMed] [Google Scholar]
- 32.Huang Y., and Liu Z. (2009) Kinetic advantage of intrinsically disordered proteins in coupled folding-binding process: a critical assessment of the “fly-casting” mechanism. J. Mol. Biol. 393, 1143–1159 [DOI] [PubMed] [Google Scholar]
- 33.Shoemaker B. A., Portman J. J., and Wolynes P. G. (2000) Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl. Acad. Sci. U.S.A. 97, 8868–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiefhaber T., Bachmann A., and Jensen K. S. (2012) Dynamics and mechanisms of coupled protein folding and binding reactions. Curr. Opin. Struct. Biol. 22, 21–29 [DOI] [PubMed] [Google Scholar]
- 35.Schwarz-Linek U., Werner J. M., Pickford A. R., Gurusiddappa S., Kim J. H., Pilka E. S., Briggs J. A., Gough T. S., Höök M., Campbell I. D., and Potts J. R. (2003) Pathogenic bacteria attach to human fibronectin through a tandem β-zipper. Nature 423, 177–181 [DOI] [PubMed] [Google Scholar]
- 36.Back C. R., Douglas S. K., Emerson J. E., Nobbs A. H., and Jenkinson H. F. (2015) Streptococcus gordonii DL1 adhesin SspB V-region mediates coaggregation via receptor polysaccharide of Actinomyces oris T14V. Mol. Oral Microbiol. 30, 411–424 [DOI] [PubMed] [Google Scholar]
- 37.Moschioni M., Pansegrau W., and Barocchi M. A. (2010) Adhesion determinants of the Streptococcus species. Microb. Biotechnol. 3, 370–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell A., Chang H. Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S., Sangrador-Vegas A., Scheremetjew M., Rato C., Yong S. Y., Bateman A., et al. (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43, D213–D221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., Heger A., Hetherington K., Holm L., Mistry J., Sonnhammer E. L., Tate J., and Punta M. (2014) Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Attwood T. K., Coletta A., Muirhead G., Pavlopoulou A., Philippou P. B., Popov I., Romá-Mateo C., Theodosiou A., and Mitchell A. L. (2012) The PRINTS database: a fine-grained protein sequence annotation and analysis resource-its status in 2012. Database 2012, bas019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sigrist C. J., de Castro E., Cerutti L., Cuche B. A., Hulo N., Bridge A., Bougueleret L., and Xenarios I. (2013) New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Servant F., Bru C., Carrère S., Courcelle E., Gouzy J., Peyruc D., and Kahn D. (2002) ProDom: automated clustering of homologous domains. Brief. Bioinform. 3, 246–251 [DOI] [PubMed] [Google Scholar]
- 43.Bryson K., Cozzetto D., and Jones D. T. (2007) Computer-assisted protein domain boundary prediction using the DomPred server. Curr. Protein Pept. Sci. 8, 181–188 [DOI] [PubMed] [Google Scholar]
- 44.Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 45.Plewczynski D., Slabinski L., Tkacz A., Kajan L., Holm L., Ginalski K., and Rychlewski L. (2007) The RPSP: Web server for prediction of signal peptides. Polymer 48, 5493–5496 [Google Scholar]
- 46.Lupas A., Van Dyke M., and Stock J. (1991) Predicting coiled coils from protein sequences. Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 47.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 49.Kiefer F., Arnold K., Künzli M., Bordoli L., and Schwede T. (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guex N., Peitsch M. C., and Schwede T. (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 30, S162–S173 [DOI] [PubMed] [Google Scholar]
- 51.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pakula R., and Walczak W. (1963) On nature of competence of transformable streptococci. J. Gen. Microbiol. 31, 125–133 [DOI] [PubMed] [Google Scholar]
- 53.Jenkinson H. F. (1987) Novobiocin-resistant mutants of Streptococcus sanguis with reduced cell hydrophobicity and defective in coaggregation. J. Gen. Microbiol. 133, 1909–1918 [DOI] [PubMed] [Google Scholar]
- 54.Edwards A. M., Potts J. R., Josefsson E., and Massey R. C. (2010) Staphylococcus aureus host cell invasion and virulence in sepsis is facilitated by the multiple repeats within FnbpA. PLoS Pathog. 6, e1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brittan J. L., and Nobbs A. H. (2015) Group B Streptococcus pili mediate adherence to salivary glycoproteins. Microbes Infect. 17, 360–368 [DOI] [PubMed] [Google Scholar]
- 56.Berrow N. S., Alderton D., Sainsbury S., Nettleship J., Assenberg R., Rahman N., Stuart D. I., and Owens R. J. (2007) A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 58.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 59.Franke D., and Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leslie A. G., and Powell H. R. (2007) in Evolving Methods for Macromolecular Crystallography (Read R. J. and Sussman J. L., eds) pp. 41–51, Springer, Netherlands [Google Scholar]
- 61.Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 62.Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 63.Evans P. R. (2011) An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 67, 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans P. R., and Murshudov G. N. (2013) How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., and Hung L.-W. (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L.-W., Read R. J., and Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baker N. A., Sept D., Joseph S., Holst M. J., and McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., and Baker N. A. (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Søndergaard C. R., Olsson M. H., Rostkowski M., and Jensen J. H. (2011) Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 7, 2284–2295 [DOI] [PubMed] [Google Scholar]
- 73.Olsson M. H., Søndergaard C. R., Rostkowski M., and Jensen J. H. (2011) PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 [DOI] [PubMed] [Google Scholar]







