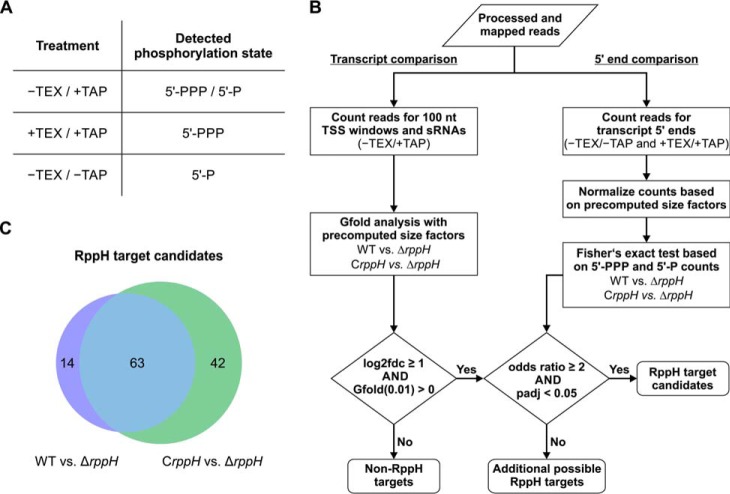
FIGURE 8.
Differential RNA-seq analysis of RNA 5′ ends in H. pylori cells containing or lacking HpRppH. A, combinations of TEX/TAP treatments used to enrich for 5′-PPP transcripts, 5′-P transcripts, or both (5′-PPP/5′-P). B, computational pipeline used to identify RppH target candidates. To pass muster, a ≥2-fold increase in both the RNA concentration (log2fdc ≥ 1) and the ratio of 5′-PPP to 5′-P ends (odds ratio ≥ 2) was required in ΔrppH cells versus WT and CrppH cells. Precomputed size factors were based on the number of mapped reads for each library. C, Venn diagram of RppH target candidates identified in ΔrppH cells versus WT or CrppH cells.