Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
HSP90 inhibition induces apoptosis in BL cells by disrupting tonic BCR signaling.
SYK is an HSP90 client protein, and BCR signaling-dependent phosphorylation of HSP90 on Y197 is required for this interaction.
Abstract
Burkitt lymphoma (BL) is an aggressive B-cell neoplasm that is currently treated by intensive chemotherapy in combination with anti-CD20 antibodies. Because of their toxicity, current treatment regimens are often not suitable for elderly patients or for patients in developing countries where BL is endemic. Targeted therapies for BL are therefore needed. In this study, we performed a compound screen in 17 BL cell lines to identify small molecule inhibitors affecting cell survival. We found that inhibitors of heat shock protein 90 (HSP90) induced apoptosis in BL cells in vitro at concentrations that did not affect normal B cells. By global proteomic and phosphoproteomic profiling, we show that, in BL, HSP90 inhibition compromises the activity of the pivotal B-cell antigen receptor (BCR)-proximal effector spleen tyrosine kinase (SYK), which we identified as an HSP90 client protein. Consistently, expression of constitutively active TEL-SYK counteracted the apoptotic effect of HSP90 inhibition. Together, our results demonstrate that HSP90 inhibition impairs BL cell survival by interfering with tonic BCR signaling, thus providing a molecular rationale for the use of HSP90 inhibitors in the treatment of BL.
Introduction
Burkitt lymphoma (BL) is an aggressive B-cell neoplasm derived from germinal center B cells and was first described in 1958 in African children, in whom it is particularly prevalent.1 Current treatment strategies for BL are based on a combination of intensive chemotherapy and anti-CD20 antibodies. The largest prospective multicenter trial for BL reported a 5-year survival rate of 84% for younger patients (<55 years), as opposed to only 62% for elderly patients (>55 years).2 In particular, elderly patients with comorbidities and patients with endemic BL in developing countries are often not eligible for intensive chemotherapy; therefore, there is a need for novel, targeted therapies.
A subset of B-cell lymphomas has been shown to depend on signals that are transduced by the B-cell antigen receptor (BCR). For instance, activated B-cell–like diffuse large B-cell lymphomas (ABC-DLBCL) depend on chronic active BCR signaling, whereas BL cells are addicted to “tonic” BCR signaling, which occurs in the absence of receptor engagement and promotes cell survival.1,3,4 A recent study systematically compared the biochemical nature of tonic and activated BCR signaling using a quantitative phosphoproteomic approach.5 This work identified BCR-dependent phosphorylation events and distinct effectors for activated and tonic BCR signaling, implying a certain degree of specificity for these 2 BCR signaling modes.5 Activated and tonic BCR signaling exhibit distinct vulnerabilities within the cell-intrinsic BCR signaling networks. In ABC-DLBCL, for example, mutated BCR signaling effectors lead to constitutively active BCR signaling and induce cell survival by activating NF-κB.6,7 In contrast, tonic BCR-dependent phosphatidylinositol 3-kinase signaling promotes the survival and proliferation of BL cells,3 whereas NF-κB activity appears to be dispensable in these cells.8,9
Despite these advances in our understanding of BCR signaling, little is known about how to target the BL-specific signaling routes for therapeutic purposes. This is particularly challenging because, in contrast to ABC-DLBCL, BL cells do not harbor genetic mutations in BCR-proximal signaling effectors.3,7,10
Here, we set out to identify vulnerabilities and their underlying mechanism in BL by combining a compound screen with quantitative phosphoproteomics and cell biology assays. We observed a strong efficacy of heat shock protein 90 (HSP90) inhibitors and found that HSP90 inhibition leads to apoptosis in BL cells by interfering with spleen tyrosine kinase (SYK)-mediated tonic BCR signaling. Taken together, these findings provide evidence for HSP90 potentially being a therapeutic target in BL.
Methods
Cells, vectors, and reagents
The lymphoma cell lines DG75, Daudi, BJAB, BL41, BL2, CA46, Namalwa, Ramos, Raji, DogKit, Gumbus, and U2932 were obtained from DSMZ (Braunschweig, Germany) or ATCC (Teddington, UK). BL7, BL60, Karpas422, and LY47 were provided by G.M. Lenoir (International Agency for Research on Cancer, Lyon, France), and Salina, Seraphine, and Cheptanges were provided by A. Rickinson, (Birmingham, United Kingdom). Cell lines were authenticated by multiplex cell authentication (Multiplexion, Heidelberg, Germany).11 All cell lines were cultured in RPMI supplemented with 10% to 20% heat-inactivated fetal bovine serum, penicillin/streptomycin, and l-glutamine (all from Invitrogen) at 37°C and 5% CO2. Stable isotope labeling with amino acids in cell culture (SILAC) of DG75 and Daudi cells was performed by culturing cells in SILAC-RPMI 1640 medium devoid of arginine and lysine (Pierce) supplemented with 10% (DG75) or 20% (Daudi) dialyzed fetal calf serum (Sigma-Aldrich, Taufkirchen, Germany), penicillin/streptomycin (Invitrogen), and the respective SILAC amino acids (all from Cambridge Isotopes, Tewksbury, MA). “Light” SILAC medium contained [12C614N4]-l-arginine and [12C614N2]-l-lysine, “medium” SILAC medium contained [13C614N4]-l-arginine and [2D4,12C6,14N2]-l-lysine, and “heavy” SILAC medium contained [13C615N4]-l-arginine and [13C615N2]-l-lysine.
For phosphoproteomic and protein expression profiling of DG75 and Daudi cells, the light-labeled cell batches were treated with dimethyl sulfoxide (DMSO), whereas the heavy-labeled cell batches were treated with the HSP90 inhibitors AT13387 or STA-9090.
Cell lysis for SILAC-labeled cells was performed as described in Oellerich et al,12 except that phosphatase inhibitor cocktails 2 and 3 (Sigma) were added according to the manufacturer’s instructions. Cell lysis for proteomic profiling of tyrosine phosphorylation was performed according to the manufacturer’s instructions by using a urea-based lysis buffer (PTMScan Kit, Cell Signaling Technology).
Compound screening and data analysis
A drug sensitivity screen was performed at 10 concentrations of 32 compounds tested (30 μM to 1.5 nM; threefold dilution). Results for 30 of these compounds are shown in Figure 1A, for which a validation in 8 of the 17 BL lines by a flow cytometric apoptosis/cell cycle analysis showed results consistent with the screening data. Screening was performed in white 384-well plates (Greiner Bio One). In each well, 10 μL of diluted compound was plated. Depending on the growth rate of the given cell line, 1500 to 7000 cells in 40 μL RPMI were added for a final working volume of 50 μL per well. Plates were sealed with breathable foils (Sigma-Aldrich) to reduce evaporation at the plate edges. After incubation for 48 hours, adenosine triphosphate levels (used as a criterion of viability) were determined by a CellTiter Glo assay (Promega). Luminescence was measured on a an Infinite F200 Microplate Reader (Tecan Group AG, Männedorf, Switzerland) with an integration time of 0.2 seconds per well. Viability was calculated as percentage of a DMSO-treated control. To exclude toxic effects of the solvent (DMSO), additional controls with PBS were performed. Dose-response curves and 50% inhibitory concentration (IC50) values were estimated using R 3.3.1 and the R package nplr (n-parameter logistic regression), version 0.1-5. A 4-parameter unweighted logistic regression model was used. Fitted curves were inspected and evaluated based on the goodness-of-fit estimator. IC50 values greater than the highest concentration (30 μM) were set to this value.
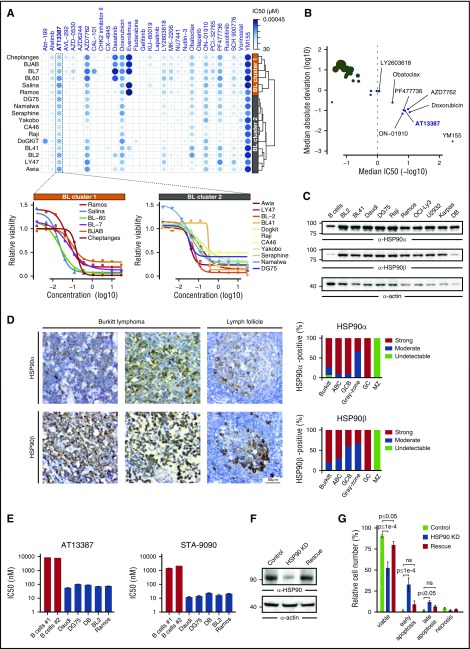
Figure 1.
HSP90 inhibition induces apoptosis of BL cells. (A, top) Heatmap of –log10 (IC50) values for the indicated compounds and BL cell lines. The hierarchical clustering of cell lines based on the corresponding drug response profiles is indicated. (Bottom) Dose-response curves for the HSP90 inhibitor AT13387 in the indicated BL cell lines. (B) Median absolute deviation (log10) of IC50 values vs median IC50 (–log10) across BL cell lines for all tested compounds. Compounds exhibiting an IC50 value <1 μM are labeled. Point sizes are proportional to median absolute deviation values. (C) Anti-HSP90α/β immunoblots of cleared cellular lysates (CCLs) from primary B cells and lymphoma cell lines. Actin was used as loading control. (D) Immunohistochemical staining of lymph node tissues from patients with BL, ABC and germinal center B-cell-like (GCB) DLBCL, gray-zone lymphoma, or healthy donors (GC, germinal center; MZ, marginal zone) with anti-HSP90α and anti-HSP90β antibodies. (Left) Representative images (40-fold magnification) are shown. (Right) All tissue sections were evaluated by 2 independent pathologists using a 3-stage staining score according to the signal intensity. (E) IC50 values for AT13387 and STA-9090 inhibitors after 48 hours of HSP90 inhibition determined by an Annexin V/7-AAD–based flow cytometric apoptosis assay. IC50 values were estimated from Annexin V/7-AAD double negative viable cells. (F) Anti-HSP90 immunoblots of CCL from Daudi cells expressing either control shRNAs, HSP90-specific shRNA, or HSP90-specific shRNA together with murine HSP90 (rescue), at day 2 after lentiviral transduction. (G) Corresponding Annexin V/7-AAD-based apoptosis readout for cells in panel F. Error bars are mean ± standard deviation (SD), n = 3. P values are from a 2-way analysis of variance test; ns, not significant.
Immunohistochemistry
Lymphoma and normal lymph node tissues were acquired from 45 patients from the University Medical Centers, Göttingen and Würzburg, Germany. Analyzed cases included 15 BLs, 10 ABC-DLBCLs, 10 germinal center B-cell–like DLBCLs, 6 gray-zone lymphomas, and 4 healthy lymph nodes. Each tissue sample was fixed in 4% buffered formalin and embedded in paraffin. Ethical approval for using the human material in the present study was obtained from the Ethics Committee of the University Medical Center Göttingen (#19-2-16). Further details are described in the supplemental Materials and methods, available on the Blood Web site.
MS and protein expression analysis
For enrichment of tyrosine-phosphorylated peptides, SILAC-labeled DG75 and Daudi cells were lysed in 8M urea buffer. The resulting proteins were mixed in equimolar amounts, digested, and then subjected to immunoprecipitation of phosphotyrosine-containing peptides using the pY1000 antibody (Cell Signaling Technology) based on the protocol described by Rush et al13 and following manufacturer’s instructions. After pY enrichment, peptides were desalted by using a self-made C18 stage-tip14 and then dried in a SpeedVac. For each sample, 2 technical replicates were analyzed by mass spectrometry (MS) as described in Corso et al.5 Protein expression profiling was performed as described previously.5
Liquid chromatography tandem MS analysis was performed on a Q Exactive HF (Thermo Fisher Scientific) mass spectrometer coupled with Ultimate 3000 RSLCnano System (Dionex). Phosphopeptides were first trapped on a homemade precolumn (ReproSil-Pur 120 C18-AQ, 5 µm; Dr. Maisch GmbH; 100 µm × 5 cm) at 10 μL/minute of loading buffer (2% acetonitrile, 0.02% trifluoroacetic acid in water) and then separated on an analytical column (ReproSil-Pur 120 C18-AQ, 1.9 µm; Dr. Maisch GmbH, 75 µm × 30 cm, self-packed) with a 90-minute linear gradient ramping from 1% to 35% of buffer B (80% acetonitrile, 0.1% formic acid) followed by another 10-minute linear gradient to 60% buffer B with corresponding composition of buffer A (0.1% formic acid in water) at a constant flow rate of 300 nl/minute. The Q Exactive HF was operated in a data-dependent acquisition mode in which 1 full MS scan across the 350 to 1600 mass-to-charge ratio (m/z) range was acquired at a resolution setting of 120 000 full width, half maximum to select up to 20 most abundant peptide precursors of charge states 2 to 6 greater than a 3 × 104 intensity threshold, at an isolation width of 1.6 m/z. Precursors were fragmented by higher collision energy dissociation with nitrogen at a normalized collision energy setting of 28%, and their product ion spectra were recorded with a start mass of 110 m/z at resolution of 30 000 full width, half maximum. Automatic gain control target value and maximum ion injection times for MS and tandem MS were 1 × 106 in 40 ms and 1 × 105 in 64 ms, respectively. Selected precursor mass-to-charge ratio values were then excluded for the following 45 seconds.
The MS raw files were processed by MaxQuant15 (version 1.5.5.1) and tandem MS spectra were searched against UniProt human database (both Swiss-Prot and TrEMBL, downloaded May 2016; 153 652 entries) via the Andromeda search engine.16 Mass tolerance after recalibration of precursor mass and fragment ion mass were set as 6 and 20 ppm, respectively. Allowed variable modifications included protein deamidation, oxidation, and phosphorylation. Cysteine carbamidomethylation was defined as a fixed modification. Minimal peptide length was set to 7 amino acids with the maximum of 2 enzymatic miscleavages. The false discovery rate was set to 1% for both peptide and protein identifications. Intensities of all identified peptides were determined by MaxQuant with enabled options “match between runs” and “requantify.” Perseus (version 1.5.5.1) was then used for downstream analyses. After removing all decoy hits and potential contaminant entries, identified phospho-sites with <.75 localization probability were discarded. All SILAC ratios were log2 transformed and those with absolute Z score >1 were defined as significantly regulated.
Pathway enrichment analysis and functional annotation
Pathway enrichment analysis was performed using the R/Bioconductor package ReactomePA, version 1.16.2 (http://bioconductor.org/packages/ReactomePA). P values were adjusted for multiple comparisons using the Benjamini-Hochberg procedure.
Immunoprecipitation experiments and immunoblotting
SYK was immunoprecipitated from 2 × 107 DG75 and Daudi cells using SYK-specific antibodies (clone D3Z1E, Cell Signaling Technology) at a 1:100 dilution, as described previously.17 In addition, HSP90 was immunoprecipitated from 2 × 107 DG75 and Daudi cells using HSP90-specific (Abcam) or hemagglutinin (HA)-specific (Roche) antibodies at a 1:200 dilution and at a concentration of 2 μg/mL, respectively. Immunoblotting was performed as described in Oellerich et al.17
Results
HSP90 inhibition leads to apoptosis in BL cells
To identify potential novel drug targets for BL treatment, we tested a panel of 30 small molecule inhibitors in 17 BL cell lines (Figure 1A). The drug response partitioned BL cell lines into 2 clusters characterized by distinct response patterns (Figure 1A). Most compounds exhibited either little or no efficacy or cell-line–specific responses (Figure 1A-B). For example, the BCL2 inhibitor Abt-199 was effective only on DoGKiT cells, which are known to harbor a bcl-2 rearrangement (t(14;18)).18 In contrast, a small set of compounds including the survivin inhibitor YM155 and the HSP90 inhibitor AT13387 strongly reduced cell viability across all tested BL lines (Figure 1A-B). Notably, the variability in the response to AT13387 across BL cell lines was comparable to that of doxorubicin, a chemotherapeutic agent commonly used for the treatment of BL (Figure 1B).
These results prompted us to further dissect the effects of Hsp90 inhibition in BL cells. First, we confirmed the expression of both constitutive (HSP90β) and inducible (HSP90α) HSP90 forms in BL cell lines and in patient-derived lymphoma samples (Figure 1C-D). We found strong HSP90α/β expression in BL and DLBCL cells, whereas gray-zone lymphomas exhibited lower HSP90 expression levels. Next, we analyzed the effects of the HSP90 inhibitors AT13387 (onalespib) and STA-9090 (ganetespib), which target both HSP90α and HSP90β, on apoptosis and cell cycle. By determining the IC50 using an Annexin V/7-AAD–based apoptosis assay, we found that BL cells were approximately 100-fold more sensitive to HSP90 inhibition (mean IC50 = 83.6 ± 24.8 nM and 18.2 ± 5.9 nM for AT13387 and STA-9090, respectively) than normal B cells purified from the peripheral blood of 2 healthy donors (mean IC50 = 9333 ± 367 nM and 1984.5 ± 386.5 nM for AT13387 and STA-9090, respectively) (Figure 1E). Moreover, HSP90 inhibition induced cell-cycle arrest in BL cells (supplemental Figure 1A). These results appear to be on-target effects of the HSP90 inhibitors because (1) a knockdown of HSP90 significantly increased apoptosis of DG75 and Daudi cells (Figure 1F-G; supplemental Figure 1B) and (2) this effect was partially rescued by reconstituting HSP90 knockdown cells with murine HSP90 (Figure 1F-G). Together, these results indicate that BL cell survival is dependent on HSP90 function.
HSP90 inhibition disrupts tonic and activated BCR signaling
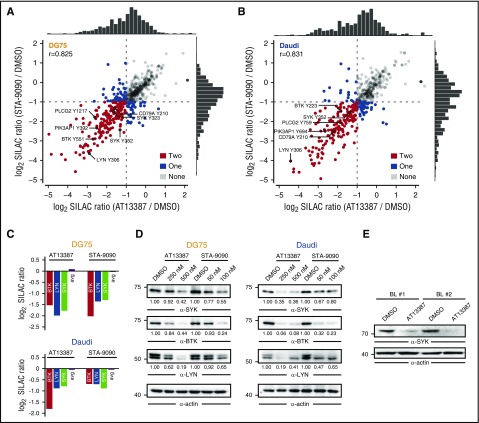
HSP90 chaperones several client proteins,19 and preclinical as well as clinical studies demonstrated the efficacy of HSP90 inhibitors in a number of cancer types.20-23 To study the effects of HSP90 inhibition at the molecular level, we analyzed the consequences of HSP90 inhibitors on the proteome- and tyrosine-enriched phosphoproteome (pYome) of BL cells using SILAC-based MS (see supplemental Figure 1C for details). We found that both AT13387 and STA-9090 induced highly correlated changes in the pYome of DG75 (r = 0.825) and Daudi cells (r = 0.831) after 24 and 16 hours of HSP90 inhibition, respectively (Figure 2A-B). Notably, these phosphoproteomic changes were also highly correlated between cell lines (r = 0.78 for AT13387; r = 0.85 for STA-9090) (supplemental Figure 1D). Pathway enrichment analysis of proteins with downregulated pY sites revealed that HSP90 inhibition mainly affects BCR-mediated processes (supplemental Figure 1E). Moreover, 45% (85/187) of the identified downregulated pY sites have been previously identified as being involved in tonic BCR signaling.5 This result suggests that HSP90 inhibition interferes with tonic antigen-independent BCR signaling. Consistently, a quantitative proteome analysis revealed downregulation of several BCR-proximal kinases including SYK, Src family tyrosine-protein kinase Lyn (LYN), and Bruton's tyrosine kinase (BTK) upon HSP90 inhibition (Figure 2C). This result was confirmed by western blotting of cleared cellular lysates (CCLs) from DG75 and Daudi cells after 24 and 16 hours of treatment with HSP90 inhibitors, respectively (Figure 2D). At these time points, >90% cell viability was retained (supplemental Figure 1F). In addition, SYK downregulation was confirmed in vitro after AT13387 treatment of primary BL cells purified from 2 patients with bone marrow involvement (Figure 2E).
Figure 2.
HSP90 inhibition attenuates tonic BCR signaling. (A-B) Scatter plot of log2 SILAC ratios normalized to DMSO for the pYome MS analysis of DG75 (A) and Daudi cells (B) after treatment with AT13387 (x-axis) or STA-9090 (y-axis). Dots are coded according to the number of HSP90 inhibitors that induced pY downregulation by a factor >2. Selected pY sites are labeled. Spearman’s rank correlation coefficients (r) are indicated and marginal distributions of SILAC ratios are shown. (C) Relative change in protein expression levels for the indicated proteins measured by SILAC-based MS after 24 hours of HSP90 inhibition. The average SILAC ratio (avg) across all detected proteins is indicated. (D) Immunoblots of CCL from DG75 and Daudi cells that were treated with DMSO, AT13387, or STA-9090 for 24 and 16 hours, respectively, using antibodies against SYK, BTK, and LYN. Actin was used as loading control. (E) Anti-SYK immunoblots of CCL from bone marrow–derived primary BL cells of 2 patients that were treated in vitro for 16 hours with either DMSO or 250 nM AT13387. Actin was used as loading control.
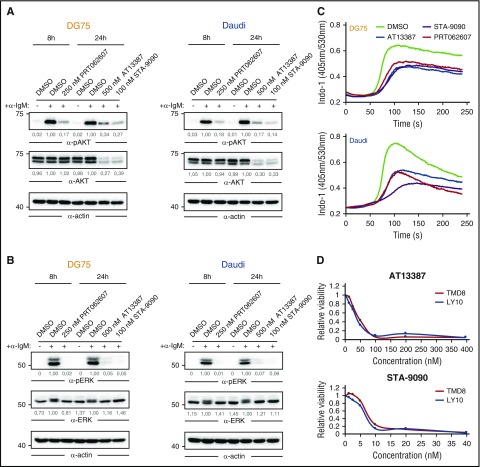
The effect of HSP90 inhibition on the pivotal BCR-proximal kinases LYN, SYK, and BTK was furthermore reflected by an attenuated BCR signaling response upon BCR stimulation. BCR-stimulated BL cells showed reduced tyrosine phosphorylation of extracellular signal regulated kinase (ERK) and AKT (Figure 3A-B) as well as reduced Ca2+-mobilization (Figure 3C) upon HSP90 inhibition. These effects were similar in cells that were treated with the SYK inhibitor PRT062607 (Figure 3A-C). Finally, we investigated the efficacy of HSP90 inhibitors on 2 ABC-DLBCL cell lines, which, in contrast to BL cells, depend on chronic active BCR signaling.7 In line with our results in BL cells, ABC-DLBCL cells exhibited reduced viability upon HSP90 inhibition (Figure 3D). Together, these results indicate that both tonic and activated BCR signaling modes are dependent on HSP90 function.
Figure 3.
HSP90 inhibition attenuates activated BCR signaling. (A-B) Immunoblots of CCL from DG75 (left) and Daudi (right) cells that were treated with DMSO, AT13387, STA-9090, or PRT062607 for indicated durations, using antibodies against (p)AKT (A) and (p) extracellular signal-regulated kinase (pERK) (B). Actin was used as loading control. (C) DG75 and Daudi cells were treated as in panels A-B, subsequently loaded with the ratiometric Ca2+ chelator INDO‐1‐AM, and subjected to Ca2+ flux analysis by flow cytometry. BCR engagement was induced after 30 seconds using 5 µg/mL goat-anti-human immunoglobulin M. (D) Cell viability of the ABC-DLBCL cell lines TMD8 and Ly10 treated with the indicated concentrations of AT13387 and STA-9090 for 24 hours. Values were determined by a 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium assay and normalized to DMSO controls.
SYK is an HSP90 client protein in BL cells
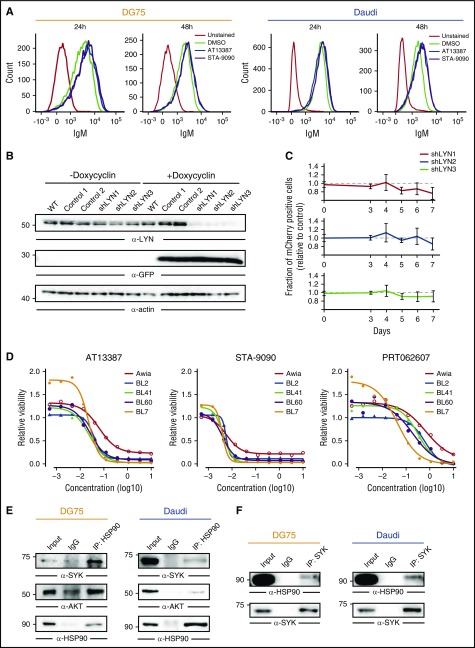
We next set out to dissect the molecular mechanism by which HSP90 inhibition attenuates BCR signaling. Flow cytometric analysis indicated that HSP90 inhibitors do not affect the expression of the BCR on the plasma membrane (Figure 4A); therefore, we focused on the BCR-proximal kinases that are downregulated upon HSP90 inhibition (Figure 2C). Neither BTK inhibition by the small molecule PCI-32765 (ibrutinib) (median IC50, 7.51 µM; see Figure 1A) nor downregulation of LYN by inducible Lyn-specific short hairpin RNAs (shRNAs) (Figure 4B-C) affected BL cell viability. In contrast, SYK inhibition negatively affected BL cell viability in a manner similar to HSP90 inhibition (Figure 4D). The congruent effects of HSP90 and SYK inhibition led us to examine whether SYK is chaperoned by HSP90. By coimmunoprecipitation followed by immunoblotting, we found that SYK and HSP90 interact at the protein level (Figure 4E-F). This result, along with reduced SYK expression upon HSP90 inhibition (Figure 2C-E), suggests that SYK is a client protein of HSP90 in BL. Notably, SYK has not been identified as an HSP90 client in other cell types, including breast cancer cells, where SYK acts as a tumor suppressor.24-26
Figure 4.
SYK is an HSP90 client protein. (A) BCR cell surface expression in DG75 and Daudi cells treated for 24 and 48 hours with either DMSO or AT13387 and STA-9090, measured by flow cytometry. Unstained cells were used as controls (gray lines). (B) Anti-LYN immunoblot of CCL from DG75 cells transduced with doxycycline-inducible constructs encoding GFP together with control shRNAs or shRNAs targeting LYN. GFP and actin served as induction and loading controls, respectively. (C) Fraction of mCherry-positive transduced cells relative to mCherry-negative untransduced cells at the indicated time points. Error bars are mean ± SD, n = 3. (D) Cell viability normalized to DMSO controls in 5 BL cell lines treated with the indicated concentrations of AT13387, STA-9090, or the SYK inhibitor PRT062607. (E-F) Coimmunoprecipitation of HSP90 and SYK. CCL from DG75 and Daudi cells were subjected to immunoprecipitation using anti-HSP90 antibodies (IP: HSP90) (E) or anti-SYK antibodies (IP: SYK) (F) or isotype-matched control antibodies (IgG) and blotted for HSP90, SYK, and AKT. CCLs are shown as input controls.
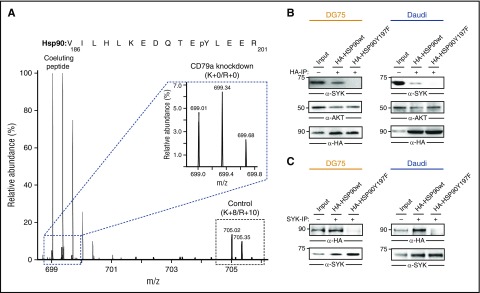
We next examined the interaction between SYK and HSP90 in BL cells. We previously found that phosphorylation of tyrosine 197 of HSP90 depends on tonic BCR signaling in BL cells (Figure 5A; supplemental Figure 2).5 Because interactions between HSP90 and several client proteins depend on posttranslational modifications of HSP90,27,28 we tested whether Y197 phosphorylation is required for the interaction between HSP90 and SYK. We addressed this question by expressing HA-tagged versions of wild-type HSP90 and a variant with a Y-to-F mutation at position 197 (HSP90Y197F) in DG75 and Daudi cells. Coimmunoprecipitation and western blot analysis revealed that the Y197 residue is necessary for HSP90–SYK interaction in BL cells (Figure 5B-C). Notably, the known HSP90 interactor AKT was purified with the Y197F variant as well, indicating a specific role of Y197 phosphorylation. Our results, therefore, not only classified SYK as an HSP90 client, but they also identified a BCR signaling-dependent phosphorylation event that is required for the chaperoning of SYK in BL cells.
Figure 5.
HSP90-pY197 is required for interaction with SYK. (A) MS spectrum showing downregulation of pY197 of HSP90 upon CD79a knockdown in DG75 cells. The inset shows the MS peaks for the light-labeled pY197-containing peptide. Here, signals from a coeluting peptide were removed for better visualization. Raw data were taken from Corso et al.5 (B) Coimmunoprecipitation of wild-type (wt) and mutant (Y197F) HA-tagged HSP90 proteins and SYK. CCL from DG75 (left) and Daudi (right) cells were subjected to immunoprecipitation using anti-HA antibodies (B) or anti-SYK antibodies (C) and blotted for HA, SYK, and AKT.
A TEL-SYK fusion protein partially rescues apoptosis induced by HSP90 inhibition
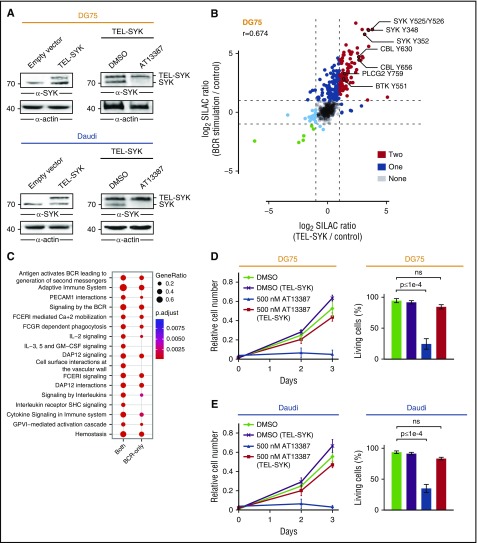
The results presented here indicate that the apoptotic effect of HSP90 inhibitors in BL cells is primarily mediated by destabilization of SYK. To further confirm this, we introduced a constitutively active TEL-SYK fusion protein in DG75 and Daudi cells.29,30 TEL-SYK expression was not significantly affected by HSP90 inhibition in these BL cell lines (Figure 6A), and it induced phosphorylation of several BCR effector proteins, including CBL, PLCγ2, and BTK. This response recapitulated the phosphorylation profile induced by BCR engagement (r = 0.674), although a subset of phosphorylation events is BCR stimulation–specific (Figure 6B-C). Furthermore, TEL-SYK expression largely rescued the apoptotic effect of the HSP90 inhibitor AT13387 in both DG75 and Daudi cells (Figure 6D-E), further supporting the functional relevance of the HSP90-SYK interaction for BL cell survival.
Figure 6.
TEL-SYK expression partially rescues the effect of HSP90 inhibition. (A) Anti-SYK immunoblot analysis of CCL from untreated DG75 and Daudi cells that were transduced with an empty control vector or a TEL-SYK fusion protein-encoding construct (left) and TEL-SYK expressing cells that were treated with DMSO or AT13387 (right). Actin was used as loading control. (B) Scatter plot of SILAC ratios normalized to untreated cells for the pYome MS analysis of DG75 cells. Untreated wild-type DG75 cells were labeled with “light” SILAC amino acids, BCR-stimulated (5 minutes) DG75 cells were labeled with “heavy” SILAC amino acids, and TEL-SYK–expressing DG75 cells were labeled with “medium” SILAC amino acids. Dots are color coded according to the number of conditions that induced pY upregulation by a factor >2. Selected pY sites are labeled. Spearman’s rank correlation coefficient (r) is indicated. (C) Pathway enrichment analysis of proteins harboring pY sites significantly regulated by both BCR stimulation and TEL-SYK expression (both) or by BCR stimulation only (BCR only). (D-E) Cell proliferation and Annexin V/7-AAD-based apoptosis assay of DG75 and Daudi cells that were transduced with an empty control vector or a TEL-SYK fusion protein-encoding construct and treated with DMSO or AT13387. Error bars are mean ± SD, n = 3. P values are from a 2-way analysis of variance test.
Discussion
Our study uncovers a hitherto unknown function for HSP90 in inducing apoptosis of BL cells by disrupting tonic BCR signaling through destabilization of the pivotal BCR transducer kinase SYK. Our findings are consistent with the previously reported HSP90 dependence of aberrantly activated signaling in solid tumor and leukemia cells.20,31
By characterizing the molecular consequences of HSP90 inhibition in BL cells, we provide several lines of evidence indicating that SYK is a client protein of HSP90. First, HSP90 inhibition strongly reduced SYK protein levels in BL cells. Second, central SYK-mediated signaling processes, including activation of the AKT pathway and mobilization of Ca2+, were markedly abrogated after HSP90 inhibition. Third, we demonstrated the physical interaction between HSP90 and SYK and its dependency on the BCR signaling-dependent phosphorylation of HSP90 on Y197. Notably, this site was previously shown to be regulated by tonic BCR signaling in BL cells.5 Moreover, our data indicate that the relevance of HSP90 inhibition extends to lymphomas other than BL. For example, HSP90 inhibition induces apoptosis in ABC-DLBCL cells that depend on chronic active BCR signaling, further expanding the scope of putative therapeutic applications.7
As expected from the broad chaperoning activity of HSP90, BCR effector kinases other than SYK, such as LYN, BTK, and AKT, become less stable after HSP90 inhibition. However, although downregulation of AKT might further support the proapoptotic effect of HSP90 inhibitors in some BL cell lines (Figure 1A), LYN and BTK are unlikely to play a major role in this process because neither BTK inhibition nor LYN downregulation by shRNA affected the survival of BL cells. The pivotal character of the SYK-HSP90 axis on BL cell survival is further emphasized by the fact that expression of a constitutively active TEL–SYK fusion protein, which is not destabilized by HSP90 inhibition, could protect BL cells from the apoptotic effect of HSP90 inhibitors.
Of importance, and in line with the assumption that the SYK-HSP90 axis is particularly relevant for the survival of BL cells, small molecule inhibitors of HSP90 are far more effective in BL cells compared with nontransformed primary B cells. This finding makes these compounds a promising therapeutic option for BL patients who are not eligible for intensive chemotherapy. The feasibility of the clinical use of HSP90 inhibitors has already been demonstrated in several clinical trials.23,32
The potential synergism between inhibitors targeting HSP90 and specific BCR-dependent kinases should be explored in future studies. Because HSP90 inhibitors target several important BCR signaling effectors at once, they might also become a valuable tool in the context of kinase inhibitor resistance, which is driven by mutations in the target kinase itself or bypass mechanisms through other cell survival promoting kinases.33-35
In summary, we identified and characterized HSP90 inhibitors as an efficient tool to induce apoptosis in BL cells, providing a basis for the development of novel and more specific therapeutic strategies for BL patients.
Acknowledgments
The authors thank Uwe Plessman, Monika Raabe, Annika Kühn, Jennifer Appelhans, Silvia Münch, and Bärbel Junge for their technical support, and Johannes Zuber for providing valuable reagents.
This work was supported by grants from the Deutsche Krebshilfe (111399) (T.O. and M.E.), and a European Molecular Biology Organization (EMBO) long-term fellowship (1305-2015 and Marie CurieActions LTFCOFUND2013/GA-2013-609409) (F.C.). Work in the Department of Haematology in Cambridge is supported by grants from Bloodwise (13003), the Wellcome Trust (104710/Z/14/Z), the Medical Research Council, the Kay Kendall Leukaemia Fund, the Cambridge National Institute for Health Research Biomedical Research Center, the Cambridge Experimental Cancer Medicine Centre, the Leukemia and Lymphoma Society of America (07037), and a core support grant from the Wellcome Trust and Medical Research Council to the Wellcome Trust Medical Research Council Cambridge Stem Cell Institute.
Footnotes
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: R.W. performed experiments/contributed to manuscript preparation; K.-T.P. performed proteomic analyses and annotated/evaluated the MS data; F.C. performed computational analyses and wrote the manuscript with T.O. and M.E.; K.T. and T.Z. performed the drug screen; C.D., U.K., L.J., R.M.Y., and M.E. performed experiments; H. Bohnenberger and A.R. performed immunohistochemistry analyses; H. Bönig provided primary cells; L.M.S. supervised experiments and provided discussion; H.S. provided discussion; H.U. supervised proteomic analyses; and T.O. designed/supervised the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas Oellerich, Department of Medicine II, Hematology/Oncology, Theodor-Stern-Kai 7, D-60590 Frankfurt am Main, Germany; e-mail: thomas.oellerich@kgu.de.
References
- 1.Schmitz R, Ceribelli M, Pittaluga S, Wright G, Staudt LM. Oncogenic mechanisms in Burkitt lymphoma. Cold Spring Harb Perspect Med. 2014;4(2):a014282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoelzer D, Walewski J, Döhner H, et al. ; German Multicenter Study Group for Adult Acute Lymphoblastic Leukemia. Improved outcome of adult Burkitt lymphoma/leukemia with rituximab and chemotherapy: report of a large prospective multicenter trial. Blood. 2014;124(26):3870-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitz R, Young RM, Ceribelli M, et al. . Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam KP, Kühn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90(6):1073-1083. [DOI] [PubMed] [Google Scholar]
- 5.Corso J, Pan KT, Walter R, et al. . Elucidation of tonic and activated B-cell receptor signaling in Burkitt’s lymphoma provides insights into regulation of cell survival. Proc Natl Acad Sci USA. 2016;113(20):5688-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenz G, Davis RE, Ngo VN, et al. . Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319(5870):1676-1679. [DOI] [PubMed] [Google Scholar]
- 7.Davis RE, Ngo VN, Lenz G, et al. . Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller U, Nilsson JA, Maclean KH, Old JB, Cleveland JL. Nfkb 1 is dispensable for Myc-induced lymphomagenesis. Oncogene. 2005;24(41):6231-6240. [DOI] [PubMed] [Google Scholar]
- 9.Klapproth K, Sander S, Marinkovic D, Baumann B, Wirth T. The IKK2/NF-kappaB pathway suppresses MYC-induced lymphomagenesis. Blood. 2009;114(12):2448-2458. [DOI] [PubMed] [Google Scholar]
- 10.Richter J, Schlesner M, Hoffmann S, et al. ; ICGC MMML-Seq Project. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44(12):1316-1320. [DOI] [PubMed] [Google Scholar]
- 11.Castro F, Dirks WG, Fähnrich S, Hotz-Wagenblatt A, Pawlita M, Schmitt M. High-throughput SNP-based authentication of human cell lines. Int J Cancer. 2013;132(2):308-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oellerich T, Grønborg M, Neumann K, Hsiao HH, Urlaub H, Wienands J. SLP-65 phosphorylation dynamics reveals a functional basis for signal integration by receptor-proximal adaptor proteins. Mol Cell Proteomics. 2009;8(7):1738-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rush J, Moritz A, Lee KA, et al. . Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23(1):94-101. [DOI] [PubMed] [Google Scholar]
- 14.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663-670. [DOI] [PubMed] [Google Scholar]
- 15.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367-1372. [DOI] [PubMed] [Google Scholar]
- 16.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794-1805. [DOI] [PubMed] [Google Scholar]
- 17.Oellerich T, Bremes V, Neumann K, et al. . The B-cell antigen receptor signals through a preformed transducer module of SLP65 and CIN85. EMBO J. 2011;30(17):3620-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiefer T, Schüler F, Knopp A, et al. . A human Burkitt’s lymphoma cell line carrying t(8;22) and t(14;18) translocations. Ann Hematol. 2007;86(11):821-830. [DOI] [PubMed] [Google Scholar]
- 19.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11(7):515-528. [DOI] [PubMed] [Google Scholar]
- 20.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10(8):537-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Modi S, Stopeck AT, Gordon MS, et al. . Combination of trastuzumab and tanespimycin (17-AAG, KOS-953) is safe and active in trastuzumab-refractory HER-2 overexpressing breast cancer: a phase I dose-escalation study. J Clin Oncol. 2007;25(34):5410-5417. [DOI] [PubMed] [Google Scholar]
- 22.Caldas-Lopes E, Cerchietti L, Ahn JH, et al. . Hsp90 inhibitor PU-H71, a multimodal inhibitor of malignancy, induces complete responses in triple-negative breast cancer models. Proc Natl Acad Sci USA. 2009;106(20):8368-8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shapiro GI, Kwak E, Dezube BJ, et al. . First-in-human phase I dose escalation study of a second-generation non-ansamycin HSP90 inhibitor, AT13387, in patients with advanced solid tumors. Clin Cancer Res. 2015;21(1):87-97. [DOI] [PubMed] [Google Scholar]
- 24.Wu Z, Moghaddas Gholami A, Kuster B. Systematic identification of the HSP90 candidate regulated proteome. Mol Cell Proteomics. 2012;11(6):M111.016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coopman PJ, Do MT, Barth M, et al. . The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature. 2000;406(6797):742-747. [DOI] [PubMed] [Google Scholar]
- 26.Taipale M, Krykbaeva I, Koeva M, et al. . Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150(5):987-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mollapour M, Tsutsumi S, Truman AW, et al. . Threonine 22 phosphorylation attenuates Hsp90 interaction with cochaperones and affects its chaperone activity. Mol Cell. 2011;41(6):672-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu W, Mollapour M, Prodromou C, et al. . Dynamic tyrosine phosphorylation modulates cycling of the HSP90-P50(CDC37)-AHA1 chaperone machine. Mol Cell. 2012;47(3):434-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuno Y, Abe A, Emi N, et al. . Constitutive kinase activation of the TEL-Syk fusion gene in myelodysplastic syndrome with t(9;12)(q22;p12). Blood. 2001;97(4):1050-1055. [DOI] [PubMed] [Google Scholar]
- 30.Kanie T, Abe A, Matsuda T, et al. . TEL-Syk fusion constitutively activates PI3-K/Akt, MAPK and JAK2-independent STAT5 signal pathways. Leukemia. 2004;18(3):548-555. [DOI] [PubMed] [Google Scholar]
- 31.Moulick K, Ahn JH, Zong H, et al. . Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90. Nat Chem Biol. 2011;7(11):818-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramalingam S, Goss G, Rosell R, et al. . A randomized phase II study of ganetespib, a heat shock protein 90 inhibitor, in combination with docetaxel in second-line therapy of advanced non-small cell lung cancer (GALAXY-1). Ann Oncol. 2015;26(8):1741-1748. [DOI] [PubMed] [Google Scholar]
- 33.Zhang SQ, Smith SM, Zhang SY, Lynn Wang Y. Mechanisms of ibrutinib resistance in chronic lymphocytic leukaemia and non-Hodgkin lymphoma. Br J Haematol. 2015;170(4):445-456. [DOI] [PubMed] [Google Scholar]
- 34.Daver N, Cortes J, Ravandi F, et al. . Secondary mutations as mediators of resistance to targeted therapy in leukemia. Blood. 2015;125(21):3236-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hertlein E, Byrd JC. Signalling to drug resistance in CLL. Best Pract Res Clin Haematol. 2010;23(1):121-131. [DOI] [PubMed] [Google Scholar]