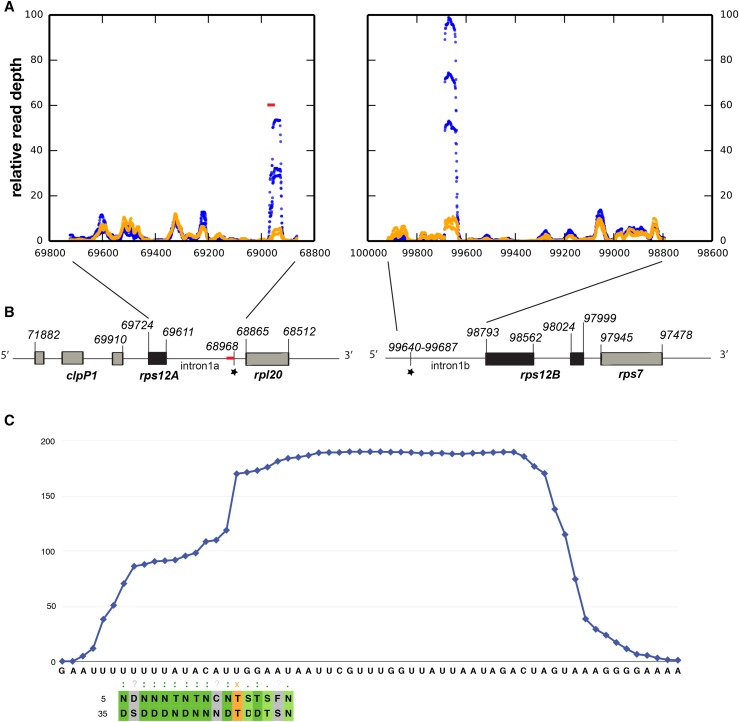
Figure 5.
RNA-seq analysis of putative “footprints” in the rps12 intron halves. A, RNA-seq was performed on gel-purified 15- to 50-nucleotide RNA fragments from partially complemented emb2654 seedlings (orange) and wild-type siblings (blue). The plots indicate the relative read depth at each nucleotide in the rps12 intron halves. Read depth has been normalized to the average depth across each intron (excluding the region of the putative footprint in each case). Data from three biological replicates are shown. The predicted EMB2654 binding site is shown by a red bar. B, Arrangement of the rps12 genes on Arabidopsis chloroplast DNA. The first exon is in rps12A, which is in the same transcription unit as rpl20 and clpP1. The second and third exons are in rps12B, located approximately 30 kb away and cotranscribed with ndhB and rps7. The position of the predicted EMB2654 binding site is indicated by a red bar, and RNA-seq footprints by black stars. Genome coordinates are indicated. C, Alignment of EMB2654 to its predicted binding site in rps12 intron 1a. The amino acids at the fifth and last positions in each PPR motif are aligned with the RNA sequence. Combinations that correlate with the aligned base (Barkan et al., 2012) are shaded in dark green. Combinations where only the fifth residue correlates with the aligned nucleotide are shaded in light green, combinations of unknown affinity are shaded in gray, and combinations that significantly anticorrelate with the aligned nucleotide are shaded in orange. The blue trace indicates the mean read depth observed in the RNA-seq analysis for wild-type samples in this region, showing the shape and extent of the footprint.