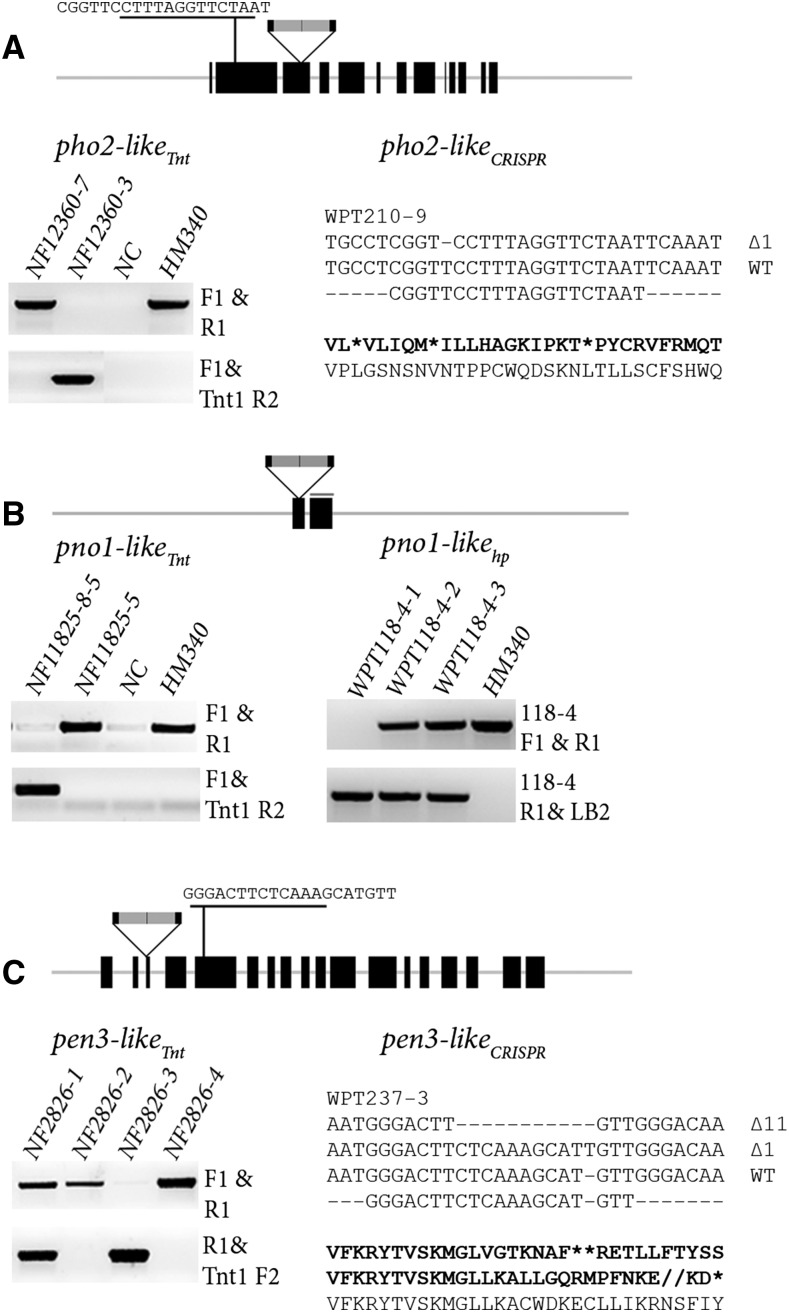
Figure 3.
Screening and identification of candidate mutant alleles. A, Two independent PHO2-like mutant alleles, pho2-likeTnt and pho2-likeCRISPR. Seeds from the Tnt1 line NF12360 were screened by PCR assay to determine homozygous and null insertions at the PHO2-like locus, identifying null insertion NF12360-7 plant and homozygous mutant NF12360-3 (Supplemental Fig. S9). A second mutant allele for PHO2-like was identified using plants transformed with a CRISPR/Cas9 reagent that targeted the open-reading frame of PHO2-like. Several homozygous T0 mutant plants were identified, and the WPT210-9 plant was selected for downstream phenotype analyses. WPT210-9 had a 1-bp deletion in the coding region of both alleles, effectively disrupting the reading frame as indicated by the three stop codons depicted in the amino acid sequence. Heritable transmission of the 1-bp mutation was confirmed by screening T1 plants by a PCR-digestion assay and sequencing. B, Two independent PNO1-like mutant alleles, pno1-likeTnt and pno1-likeHP. Tnt1 mutant alleles were identified for the PNO1-like gene by screening seed from the NF11825 line. Pno1-like mutant and null insertion lines were identified: NF11825-8 and NF11825-5, respectively. Since an insufficient number of seed was recovered from the NF11825-8 plant for phenotype analysis, seed from the next generation (NF11825-8-5) was used as wild-type plant. For the second mutant allele, a hairpin line was identified. TAIL-PCR was used to identify the genomic location of the transgene, and a PCR assay using both the genome location and transgene specific primers was used to identify a plant homozygous for the hairpin transgene (WPT118-4-1). C, Two independent PEN3-like mutant alleles, pen3-likeTnt and pen3-likeCRISPR. Seed from the NF2826 Tnt1 line were used to identify homozygous (NF2826-3) and null (NF2826-2) insertion mutant plants using both PEN3-like and Tnt1 specific primers. For the second mutant allele, we identified several CRISPR/Cas9 homozygous T0 mutant plants such as WPT237-3, which had a segregating a biallelic mutation consisting of a 11-bp deletion and a 1-bp insertion. Screening of WPT237-3 T1 mutant plants confirmed heritable transmission of both mutations.