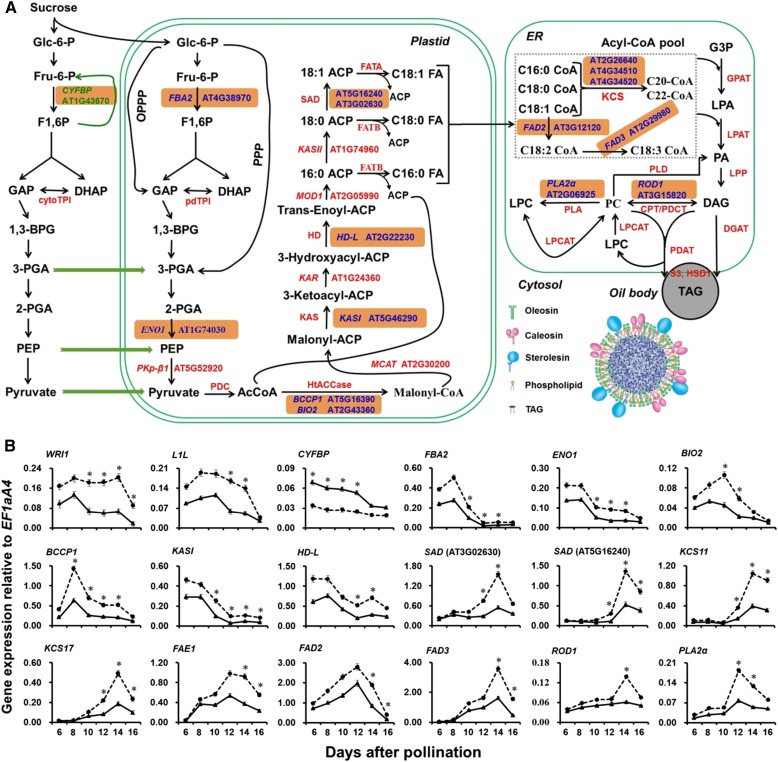
Figure 4.
Dynamic expression analysis of genes involved in the processes of glycolysis, FA biosynthesis and modification, and TAG accumulation in developing seeds of wild-type (Col-0) and myb89-1 plants. A, Simplified scheme showing the altered expression levels of genes involved in the oil biosynthetic pathway in developing seeds between wild-type (Col-0) and myb89-1 plants. The seed oil body generated from the endoplasmic reticulum (ER) accumulates TAG inside and is surrounded by a layer of phospholipids and proteins, including oleosin, caleosin, and steroleosin. The genes marked by blue color with an orange box were significantly up-regulated, and the gene CYFBP indicated by green color with an orange box was down-regulated in myb89-1 developing seeds. F1,6P, Fru-1,6-diphosphate; GAP, glyceraldehyde-3-phosphate; DHAP, dihydroxy-acetone-phosphate; cytoTPI, cytosolic triose phosphate isomerase; pdTPI, plastid TPI; 1,3-BPG, 1,3-bisphosphoglycerate; 3PGA, glycerate-3-phosphate; 2PGA, glycerate-2-phosphate; PEP, phosphoenolpyruvate; PDC, pyruvate dehydrogenase complex; AcCoA, acetyl-CoA; HtACCase, heteromeric acetyl-CoA carboxylase; ACP, acyl carrier protein; KAS, 3-ketoacyl-ACP synthase; HD, hydroxyacyl-ACP dehydratase; SAD, stearoyl-ACP desaturase; FAT, fatty acyl-ACP thioesterase; KCS, 3-ketoacyl-CoA synthase; G3P, glycerol 3-phosphate; GPAT, G3P acyltransferase; LPA, lysophosphatidic acid; LPAT, LPA acyltransferase; PA, phosphatidic acid; LPP, lipid phosphate phosphatase; DAG, diacylglycerol; PC, phosphatidylcholine; PDCT, PC:DAG phosphocholine transferase; CPT, choline-phosphotransferase; PLA, phospholipase A; PLD, phospholipase D; LPCAT, lyso-PC acyltransferase; LPC, lyso-PC; DGAT, DAG acyltransferase; PDAT, phospholipid:DAG acyltransferase; TAG, triacylglycerol; HSD, hydroxysteroid dehydrogenase; CYFBP, CYTOSOL FRU-1,6-BISPHOSPHATASE; FBA2, FRU-1,6-BISPHOSPHATE ALDOLASE2; ENO1, PLASTID-LOCALIZED PHOSPHOENOLPYRUVATE ENOLASE1; PKp-β1, PLASTIDIC PYRUVATE KINASE β-SUBUNIT1; BCCP1, BIOTIN CARBOXYL-CARRIER PROTEIN1; BIO2, BIOTIN AUXOTROPH2; MCAT, MALONYL-COA:ACP TRANSACYLASE; KAR, 3-KETOACYL-ACP REDUCTASE; MOD1, MOSAIC DEATH1; FAD2, FATTY ACID DESATURASE2; ROD1, REDUCED OLEATE DESATURATION1; PLA2α, PHOSPHOLIPASE A2α; OPPP, oxidative pentose phosphate pathway; PPP, nonoxidative pentose phosphate pathway. B, qRT-PCR analysis of the expression of genes involved in the oil biosynthetic pathway in developing seeds between wild-type (Col-0) and myb89-1 plants. Solid and dotted lines indicate the dynamic expression of genes in wild-type and myb89-1 plants, respectively. Results were normalized against the expression of EF1aA4 as an internal control. Values are means ± sd (n = 3). Asterisks indicate significant differences in gene expression levels in myb89-1 plants compared with those in wild-type plants (two-tailed paired Student’s t test, P ≤ 0.05).