The serine/threonine protein phosphatase SLP2 is mitochondrial intermembrane space localized and redox activated by the oxidoreductase Mia40.
Abstract
Reversible protein phosphorylation catalyzed by protein kinases and phosphatases represents the most prolific and well-characterized posttranslational modification known. Here, we demonstrate that Arabidopsis (Arabidopsis thaliana) Shewanella-like protein phosphatase 2 (AtSLP2) is a bona fide Ser/Thr protein phosphatase that is targeted to the mitochondrial intermembrane space (IMS) where it interacts with the mitochondrial oxidoreductase import and assembly protein 40 (AtMIA40), forming a protein complex. Interaction with AtMIA40 is necessary for the phosphatase activity of AtSLP2 and is dependent on the formation of disulfide bridges on AtSLP2. Furthermore, by utilizing atslp2 null mutant, AtSLP2 complemented and AtSLP2 overexpressing plants, we identify a function for the AtSLP2-AtMIA40 complex in negatively regulating gibberellic acid-related processes during seed germination. Results presented here characterize a mitochondrial IMS-localized protein phosphatase identified in photosynthetic eukaryotes as well as a protein phosphatase target of the highly conserved eukaryotic MIA40 IMS oxidoreductase.
Reversible protein phosphorylation is a key regulatory mechanism controlling nearly all biological processes, with at least 75% of all eukaryotic proteins suggested to be phosphorylated (Sharma et al., 2014). This regulatory mechanism is governed by the opposing actions of protein kinases and protein phosphatases. In humans and Arabidopsis (Arabidopsis thaliana) the canonical eukaryotic PPP Ser/Thr protein phosphatase complement is encoded by 13 and 26 catalytic subunits respectively, most of which require interaction with regulatory proteins to direct their specificity within the cell (Moorhead et al., 2009; Heroes et al., 2013; Uhrig et al., 2013b). In Arabidopsis and all other plants, we have uncovered a unique subfamily of prokaryotic-like PPP-family phosphatases called the Shewanella-like protein (SLP) phosphatases (Andreeva and Kutuzov, 2004; Uhrig and Moorhead, 2011; Uhrig et al., 2013a), which was found to further divide into two distinct groups called SLP1 and SLP2, differing in both subcellular localization and spatial expression across the plant (Uhrig and Moorhead, 2011). Characterization of SLP phosphatases from Arabidopsis (At) showed that AtSLP1 is chloroplast targeted and expressed exclusively in photosynthetic tissues, while AtSLP2 is likely cytosolic and highly expressed in nonphotosynthetic tissues (Uhrig and Moorhead, 2011).
With 95% of mitochondrial targeted proteins being nuclear encoded and in need of import from the cytosol, a diversity of protein import mechanisms have evolved across eukaryotes (Hentchel and Escalante-Semerena, 2015). Of recent interest is a unique mitochondrial targeting peptide independent protein import mechanism responsible for the import and assembly of mitochondrial intermembrane space (IMS) proteins. This mechanism consists of two proteins, Essential for Respiration and Vegetative Growth 1 and Mitochondrial Import and Assembly 40 (ERV1, MIA40; Herrmann and Riemer, 2012). These two proteins form a redox relay with cytochrome c, with MIA40 being responsible for catalyzing disulfide bridge formation on target proteins, trapping them in the IMS (Banci et al., 2009; Sideris et al., 2009).
Seed germination is a process largely governed by the opposing effects of two key phytohormones, abscisic acid (ABA) and gibberellic acid (GA; Kucera et al., 2005). ABA functions to establish dormancy in developing seeds while maintaining a negative regulatory role during seed germination by preserving seed dormancy (Müller et al., 2006). Conversely, GA positively regulates seed germination by stimulating seed swelling, endosperm rupture, and testa cracking to facilitate radicle emergence and seedling establishment (Holdsworth et al., 2008; Piskurewicz et al., 2008). Emphasizing the interdependence of ABA and GA in seed germinative processes is the GA biosynthetic mutant ga1, which also exhibits enhanced dormancy and requires exogenous GA to germinate, while DELLA transcription factor signaling mutant rgl2 demonstrates reduced seed dormancy resulting from enhanced GA-induced gene expression (Lee et al., 2002; Cao et al., 2005; Kucera et al., 2005). A loss of both GA biosynthesis (ga1-3) and signaling through crosses with higher order rga1/rgl2/gai mutants results in GA-independent germination (Cao et al., 2005).
Through a combination of biochemistry, cell biology, and pharmacological data, the involvement of a Ser/Thr protein phosphatase in regulating GA-related processes during seed germination has been shown. AtSLP2 was found to uniquely reside in the mitochondrial IMS. Here, it specifically interacts with mitochondrial IMS import and assembly protein 40 (AtMIA40) yielding intramolecular disulfide bonds that activate the protein phosphatase to negatively regulate seed germination by inhibiting GA related processes.
RESULTS
AtSLP2 Interacts with Mitochondrial Redox Relay Protein AtMIA40
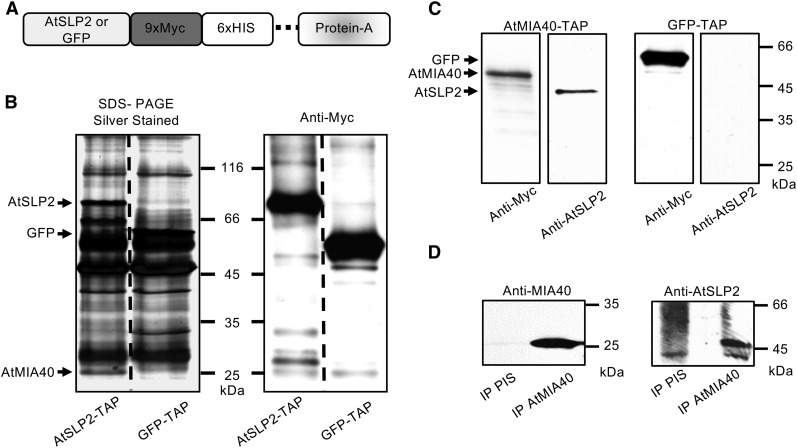
AtSLP2 was found to be specifically expressed in dark-grown Arabidopsis cell culture, roots, and seeds, while its paralog AtSLP1 was not found in any of these tissues but exclusively in photosynthetic tissue types (Supplemental Fig. S1; Uhrig and Moorhead, 2011). In order to identify potential protein interactors of AtSLP2, Arabidopsis roots and cell culture constitutively expressing tandem affinity purification (TAP)-tagged AtSLP2 (35Spro::AtSLP2-TAP; AtSLP2-TAP) were used (Fig. 1A). AtSLP2-TAP was overexpressed in both wild-type Arabidopsis cell culture and plants alongside controls GFP-TAP (35Spro::GFP-TAP) in Arabidopsis cell culture and AtSLP1-TAP (35Spro::AtSLP1-TAP) in Arabidopsis plants and cell culture. AtSLP2- and control-TAP pull-downs were performed in parallel, with each isolated TAP-tagged protein detected by immunoblot using anti-Myc antibodies (Fig. 1B). Figure 1B depicts the typical AtSLP2- and GFP-TAP interactome isolated using TAP constructs stably expressed in cell culture. The highly enriched ∼25-kD polypeptide in the AtSLP2-TAP lane was excised and identified by mass spectrometry as Cox19-like CHCH family MIA40 protein (AtMIA40). Peptide coverage was 57.4%, and no AtMIA40 peptides were recovered in a digest of the equivalent gel piece in the control lane. In parallel, a subtractive mass spectrometry approach involving the direct comparison of on-bead digested AtSLP2-, AtSLP1-, and control-TAP pull-downs was performed to identify additional AtSLP2 protein interactors (Supplemental Table S1). AtSLP2-specific binding partners found in both dark-grown Arabidopsis cell culture and roots included AtMIA40. Other potential interacting proteins, although enriched compared to the control-TAP pull-downs, do not reside in the same compartment as AtSLP2 and were also recovered with the SLP1-TAP, which is chloroplastic, suggesting they are likely not real AtSLP2 partners. For this reason, we chose to explore the association of AtSLP2 with AtMIA40.
Figure 1.
AtSLP2-TAP pull-downs isolate AtSLP2-specific protein interactors. A, Cartoon depiction of the C-terminal TAP tag in frame with either AtSLP2 or GFP. The dashed line represents a human rhinovirus 3C protease cleavage site. B, Representative silver-stained SDS-PAGE and anti-Myc immunoblot of TAP pull-downs from stably transfected dark-grown Arabidopsis cell culture. GFP-TAP and AtSLP1-TAP pull-downs offered a control for nonspecific protein interactors (Supplemental Table S1 and raw data deposited at massive.ucsd.edu). Lanes contain 30 µL (silver) and 5 µL (western) of TAP-purified protein from 50 g of dark-grown Arabidopsis cell culture. C, Reciprocal AtMIA40-TAP pull-downs verify specific interaction between AtSLP2 and AtMIA40. AtMIA40-TAP was stably transfected into dark-grown Arabidopsis cell culture and used as bait to pull-down endogenous partners, which included AtSLP2. AtSLP2 was absent from in-parallel GFP-TAP pull-downs. D, Immunoprecipitation of endogenous AtMIA40-AtSLP2 protein complex using affinity-purified anti-AtMIA40 IgG and preimmune serum IgG (PIS). Immunoblotting was performed using 0.5 µg/mL anti-Myc (ICL), 1:1,000 anti-AtMIA40, and 1.2 µg/mL affinity-purified anti-AtSLP2 (Uhrig and Moorhead, 2011).
Verification of a specific AtSLP2-AtMIA40 protein complex was supported through reciprocal TAP pull-downs using AtMIA40-TAP (35Spro::AtMIA40-TAP) stably expressed in dark-grown Arabidopsis cell culture (Fig. 1C). Control-TAP pull-downs from the same tissue failed to retrieve AtMIA40, corroborating the specificity of the observed AtSLP2-AtMIA40 interaction (Supplemental Table S1). Further support for an AtSLP2-AtMIA40 protein complex was observed by coimmunoprecipitation of endogenous AtSLP2 and AtMIA40 using affinity-purified anti-AtMIA40 IgG (Fig. 1D; Supplemental Figs. S2–S4).
AtSLP2 and AtMIA40 Colocalize to Mitochondria
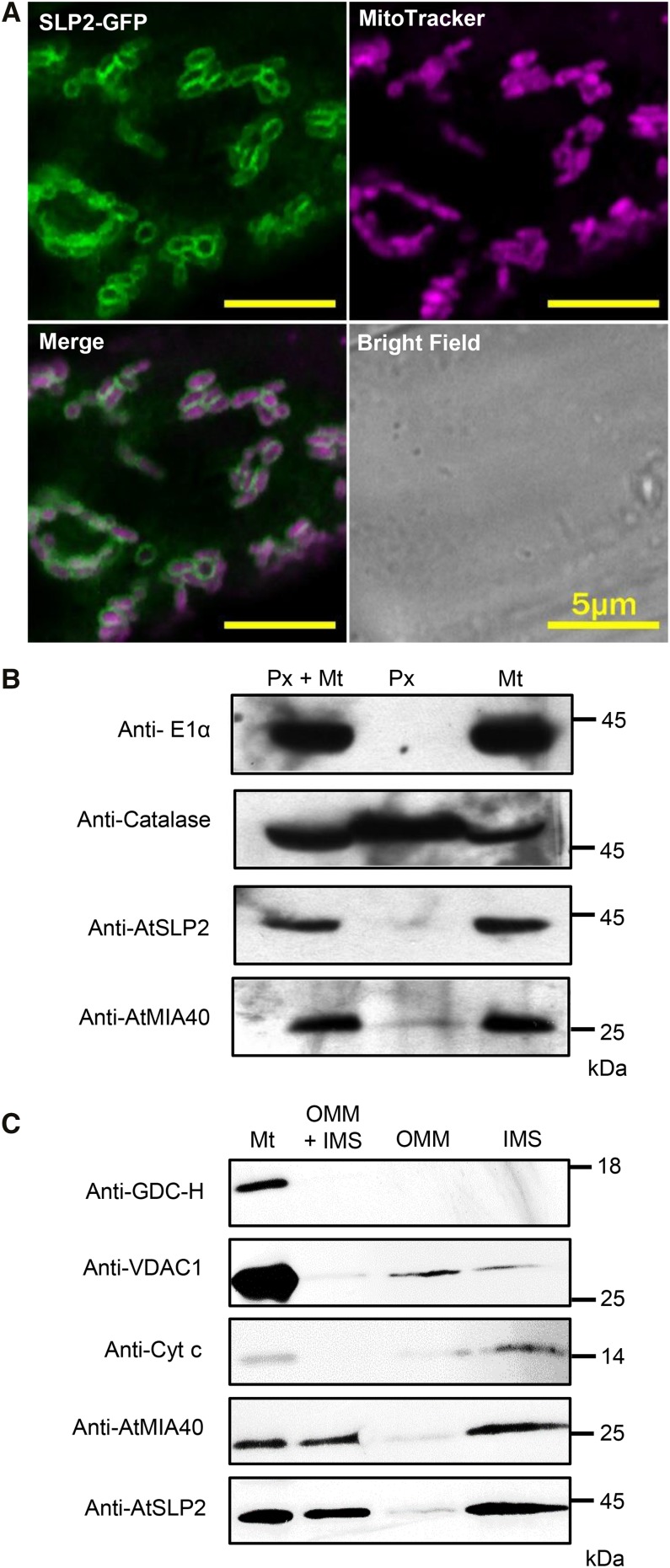
As in other eukaryotes, Arabidopsis MIA40 is a mitochondrially targeted IMS oxidoreductase (Hentchel and Escalante-Semerena, 2015). Utilizing plants stably overexpressing AtSLP2-GFP, we found colocalization with MitoTracker Deep Red FM fluorescent staining, which specifically visualizes mitochondria (Fig. 2A; Supplemental Fig. S5A). Consistent with this finding, overlapping signals from Arabidopsis stably expressing AtSLP2-GFP (35Spro:AtSLP2-GFP) and AtMIA40-RFP (35Spro:AtMIA40-RFP) confirmed AtSLP2 colocalization with AtMIA40 in the mitochondria (Supplemental Fig. S5B). Plant MIA40 has been reported to also reside in peroxisomes (Hell, 2008; Banci et al., 2009; Sideris et al., 2009; Herrmann and Riemer, 2012; Hentchel and Escalante-Semerena, 2015). Using Vicia faba leaves transiently expressing a peroxisome-targeted GFP and AtSLP2-RFP, AtSLP2 was demonstrated to not be peroxisomal (Supplemental Fig. S5C). As a further control, V. faba plants transiently expressing mitochondrial-targeted GFP (Mito-GFP; Supplemental Fig. S5, D–F) determined that within the MitoTracker Deep Red FM channel, chloroplasts autofluoresce, but that the Deep Red FM dye is mitochondrial specific.
Figure 2.
AtSLP2 and AtMIA40 are located in the mitochondrial IMS. A, Stably transfected Arabidopsis roots expressing 35Spro::AtSLP2-GFP (green) are stained with MitoTracker Deep Red FM (purple) to illuminate mitochondria. Representative individual, merged, and bright field images are shown. B, Intact mitochondria and peroxisomes were isolated from dark-grown wild-type Arabidopsis cell culture. Immunoblotting of the purified mitochondrial (Mt) and peroxisomal (Px) fractions were performed using 1:100 antimitochondrial pyruvate dehydrogenase complex E1α (anti-E1α; mitochondrial matrix) and 1:200 anticatalase (peroxisome) IgG. Each lane contains 15 µg of total protein. C, Isolated mitochondria (Mt) were further subfractionated into OMM and IMS and immunoblotted using 1:5000 anti-H protein of Gly decarboxylase complex (anti-GDC-H; mitochondria matrix marker), 1:5,000 antivoltage-dependent anion-selective channel protein 1 (anti-VDAC1; OMM marker) and 2 µg/mL anticytochrome c (anticyt c; IMS marker) IgG. Each lane contains 10 µg of total protein. Immunoblotting was performed in conjunction with affinity-purified anti-AtSLP2 IgG (Uhrig and Moorhead, 2011) and anti-AtMIA40 crude immune serum (Supplemental Fig. S4) used at 1.2 µg/mL and a 1:1,000 dilution, respectively.
To support our imaging data, we isolated intact mitochondria and peroxisomes from dark-grown wild-type Arabidopsis cell culture (Fig. 2B). Immunoblotting with AtMIA40- (Supplemental Figs. S3 and S4) and AtSLP2-specific antibodies demonstrated that endogenous AtMIA40 resides in both organelles, while AtSLP2 is mitochondrion specific (Fig. 2B). Additional immunoblotting of both the purified mitochondria and peroxisome fractions with antibodies to mitochondrial pyruvate dehydrogenase complex protein E1α (Szurmak et al., 2003) confirmed that the peroxisomal fraction was completely free of mitochondria. Given that AtMIA40 proteins reside in the IMS (Hentchel and Escalante-Semerena, 2015), we further fractionated isolated mitochondria into IMS and outer mitochondrial membrane (OMM) and looked for AtSLP2 localization (Fig. 2C). Consistent with forming a complex, both AtMIA40 and AtSLP2 were highly enriched in the IMS fraction with the IMS marker cytochrome c.
AtMIA40 Activates the Ser/Thr Phosphatase Activity of AtSLP2 through Disulfide Bond Formation
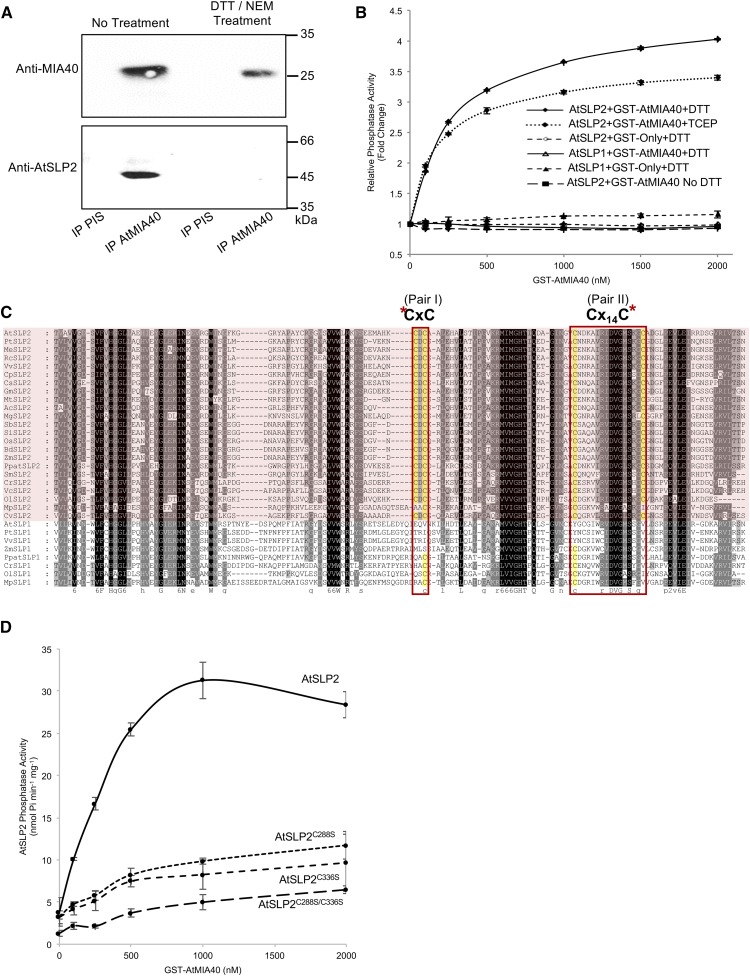
MIA40 proteins are oxidoreductases that aid the formation of disulfides on target proteins by first forming intermolecular disulfides between MIA40 and its partner (Banci et al., 2009). To determine if AtMIA40 associates with AtSLP2 in the absence of disulfide bonds, we performed anti-AtMIA40 immunoprecipitations using extracts treated sequentially with or without dithiothreitol (DTT) and the irreversible thiol-modifying agent N-ethylmaleimide (NEM). This revealed that the association of AtMIA40 with AtSLP2 is dependent upon disulfide bond formation (Fig. 3A). Activity assays employing the phosphatase substrate pNPP and recombinant AtSLP2 exhibited an AtMIA40 concentration-dependent increase in phosphatase activity (Fig. 3B) and that activation of HIS6-AtSLP2 by GST-AtMIA40 was dependent on the presence of reductant (Fig. 3B). Importantly, GST-AtMIA40 did not activate the AtSLP2 paralog, AtSLP1.
Figure 3.
AtSLP2 is redox activated by AtMIA40. A, Immunoprecipitation of endogenous AtSLP2-AtMIA40 complex is redox dependent. Treatment involved sequential reduction (20 mm DTT) and alkylation (110 mm NEM) of a dark-grown wild-type Arabidopsis cell culture extract prior to immunoprecipitation (PIS, preimmune serum IgG). B, HIS6-AtSLP2 (250 ng) incubated with increasing amounts of GST-AtMIA40 in the presence of 5 mm DTT or TCEP activates AtSLP2, while failing to activate AtSLP1. Fold-change in AtSLP2 activity was derived from AtSLP2 assays without AtMIA40. C, Alignment of SLP1 and SLP2 (pink shading) orthologs from across photosynthetic eukaryotes revealed conserved SLP2 Cys pairs. Alignment was created using MAFFT (http://mafft.cbrc.jp/alignment/software/). Asterisks denote cysteines (yellow) targeted for site-directed mutagenesis. Protein name, gene identifier, organism and sequences are presented in Supplemental Table S4. D, Site-directed mutagenesis of identified SLP2-specific cysteines in AtSLP2 results in reduced AtSLP2 activation by AtMIA40. Error bars represent ± se (n = 3).
Yeast and human MIA40s contain a specific CPC Cys pair that forms sequential disulfide bonds on its partner protein to yield the mature oxidized, properly folded IMS protein (Mordas and Tokatlidis, 2015). To date, MIA40 proteins have been observed to form two disulfide bonds on Cys pairs of client proteins that are typically separated by three or nine amino acids, although many variants of this spacing pattern have recently emerged (Mordas and Tokatlidis, 2015). Given that AtSLP1 cannot be activated by AtMIA40, we aligned AtSLP1 with AtSLP2 orthologs and searched for unique Cys pairs in AtSLP2 that may form disulfides. This analysis revealed CxC and Cx14C pairs unique to SLP2 phosphatases (Fig. 3C). Individual mutation of one Cys from each pair (single mutant) or concurrent mutation of one Cys from each pair (double mutant) almost completely abolished AtSLP2 activation by AtMIA40, indicating these conserved Cys pairs form disulfide bridges to mature AtSLP2 in the IMS (Fig. 3D; Supplemental Fig. S6).
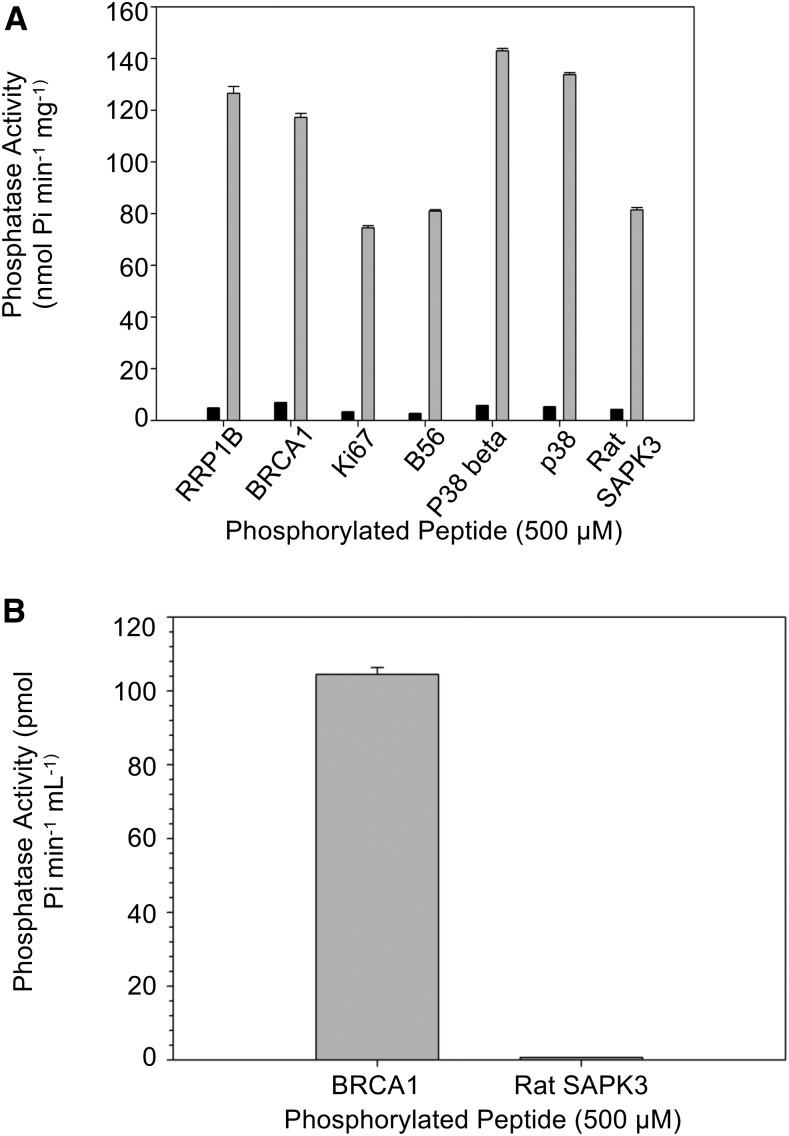
Bacterially expressed and purified HIS6-AtSLP2 was also found to possess an AtMIA40-dependent increase in activity toward pSer, pThr, pTyr and pThr/pTyr phosphorylated peptides (Fig. 4A; Supplemental Table S2). Despite an almost 20-fold activation in HIS6-AtSLP2 activity, AtMIA40 did not alter the specificity of AtSLP2 toward any one substrate peptide (Fig. 4A) and also suggested that AtSLP2 was a potential dual specificity protein phosphatase. However, assays conducted using AtSLP2-TAP isolated from dark-grown Arabidopsis cell culture showed that AtSLP2 displayed a clear preference for a pThr-containing peptide substrate over a pTyr-containing peptide (Fig. 4B). Collectively, these results indicate that AtMIA40 functions to dramatically enhance AtSLP2 activity, but not alter its phosphoprotein specificity. Furthermore, data indicate that this is accomplished through the formation of AtMIA40 catalyzed disulfide bridges as previously described for other MIA40 orthologs (Banci et al., 2009; Sideris et al., 2009).
Figure 4.
Substrate specificity of AtSLP2-AtMIA40 complex. A, A panel of phospho-peptides was screened using 250 ng of bacterially expressed and purified HIS6-AtSLP2 with (gray bars) and without (black bars) the addition of HIS6-AtMIA40 to assess the influence of AtMIA40 on the substrate specificity of AtSLP2. Phospho-peptides depicted here represent pThr (RRP1B, BRCA1), pSer (Ki67, B56), pThr/pTyr (p38, p38beta) and pTyr (Rat SAPK3), containing peptides. B, AtSLP2-TAP is pSer/pThr specific. AtSLP2-TAP activity was assessed using BRCA1 (pThr) and Rat SAPK3 (pTyr) peptide substrates. Maximal AtSLP2-TAP activity was achieved using exogenous HIS6-AtMIA40. All assays were conducted at 30°C for 1 h, with enzymatic activity deduced with malachite green. Error bars represent ± se (n = 3).
AtSLP2 and AtMIA40 Function to Negatively Regulate Seed Germination
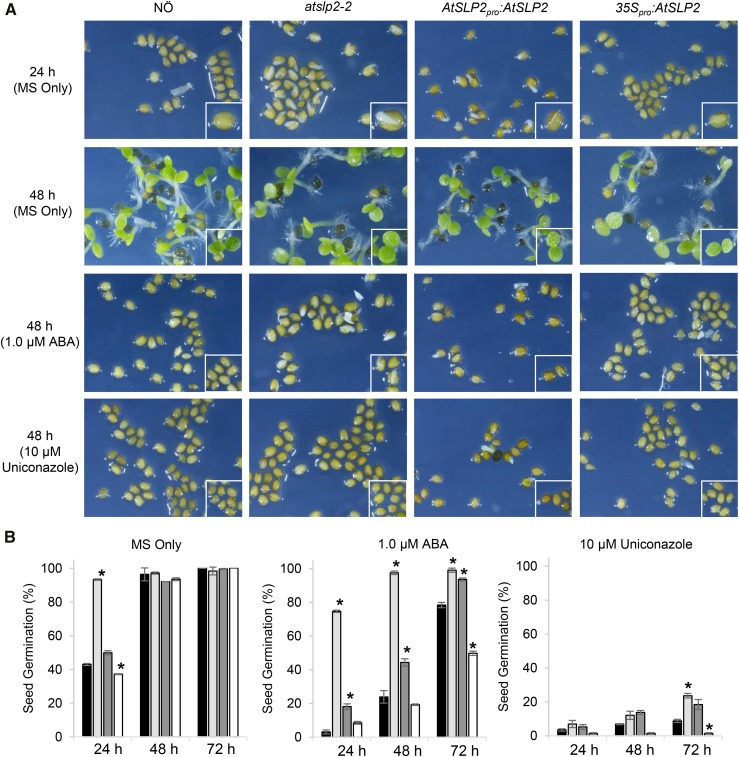
To elucidate the biological function of the protein phosphatase AtSLP2, an insertional knockout (atslp2-2) mutant was screened for growth phenotypes. Despite the availability of other atslp2 insertional mutant plants, exhaustive screening failed to identify a second knockout where AtSLP2 protein levels were diminished. This lead us to complement atslp2-2 using an endogenous promoter-driven AtSLP2 construct (AtSLP2pro::AtSLP2), in addition to generating constitutive AtSLP2 (35Spro::AtSLP2) overexpression plant lines. As shown in Supplemental Figure S7, western blotting confirmed atslp2-2 as a complete knockout, which could be complemented (AtSLP2pro::AtSLP2). Examination of atslp2-2 seedlings found they displayed accelerated germination, while 35Spro::AtSLP2 seedlings were delayed in germination (Fig. 5). Complementation of atslp2-2 with AtSLP2pro::AtSLP2 rescued the early germination phenotype.
Figure 5.
Analysis of seed germination on 0.5× MS-agar plates containing either ABA or Uniconazole. Control plates consisted of 0.5× MS only. A, Seed germination exhibited by wild-type NÖ, atslp2-2, and AtSLP2pro::AtSLP2 complemented atslp2-2 and 35Spro::AtSLP2 seeds. B, Quantitative analysis of AtSLP2 seed germination. Plant lines wild-type NÖ (black; far left), atslp2-2 (light gray; middle left); AtSLP2pro::AtSLP2 complemented atslp2-2 (dark gray, middle right) and 35Spro::AtSLP2 seeds (white; right). Application of 1.0 µm ABA slowed, but did not abolish, seed germination in atslp2-2 seeds, while nearly all seeds were equally inhibited by the application of 10 µm Uniconazole. A seed germination time-course examining multiple ABA and Uniconazole concentrations over 72 h is also shown (Supplemental Fig. S10). All seeds were imbibed at 4°C for 48 h on each plate prior to germination under 16 h light:8 h dark at 23°C. Error bars represent ± se (n = 3 plates; average of 80–120 seeds per plate). Asterisks denote two-tailed Student’s t test (P < 0.05) when atslp2-2, AtSLP2pro::AtSLP2, and 35Spro::AtSLP2 seeds were compared to wild-type NÖ. F-test indicated normal data distribution.
Further examination of AtSLP2 and AtMIA40 protein levels during imbibition and germination was performed (Supplemental Fig. S8). Here “seed imbibition” refers to seed water uptake and swelling at 4°C in the dark, while “seed germination” refers to seeds exposed to 24 h light at room temperature after imbibition. These two processes were explicitly separated to fully resolve when AtSLP2 and AtMIA40 protein levels were maximal (Supplemental Fig. S8). AtSLP2 and AtMIA40 protein levels were found to largely parallel each other over the course of imbibition and germination with protein levels peaking early in imbibition (3–6 h).
AtSLP2-AtMIA40 Protein Complex Negatively Regulates GA Biosynthesis
The observed early germination phenotype in atslp2-2 was suggestive of an alternation in hormone-related processes. As seed germination is governed by complex hormone-linked metabolism and signaling mechanisms centering around the antagonistic interaction displayed by ABA and GA, enhanced GA biosynthesis/signaling or ABA insensitivity could lead to rapid germination. To explore this hypothesis, a pharmacological approach was undertaken to test the ability of atslp2-2 to exhibit accelerated germination phenotypes in the presence of either Uniconazole (GA biosynthetic inhibitor) or ABA (to ascertain ABA insensitivity). We performed germination assays over a range of Uniconazole (0–100 µm) and ABA (0–5 µm) concentrations (Supplemental Fig. S9) with a detailed analysis of the effect of 10 µm and 1 µm Uniconazole and ABA, respectively (Fig. 5). Uniconazole (10 µm) almost completely inhibited the observed germination phenotype of atslp2-2, while 1 µm ABA delayed, but did not inhibit, atslp2-2 germination (Fig. 5). These observations implicate endogenous GA levels as responsible for the observed germination phenotype of atslp2-2, suggesting that there are increased GA levels in the absence of AtSLP2.
We further analyzed publicly available microarray data and found that AtSLP2 transcripts are down-regulated in response to ABA (Supplemental Fig. S10A) and up-regulated in response to GA (Supplemental Fig. S10B) during seed imbibition. GA-induced expression of AtSLP2 negatively correlated with Arabidopsis GA3 oxidase (GA3OX; At5g25900) and GID1A (At3g05120), while positively correlating with DELLA transcription factors RGA1 (At1g14920) and RGL2 (At3g03450) that act downstream of GA production. Furthermore, wild-type Arabidopsis seeds germinated under control conditions revealed that AtSLP2 transcript maintains an in-parallel decrease in expression with GA3OX, GID1A, RGL2, and RGA1 (Supplemental Fig. S11A), while also being slightly down-regulated in germinating seeds grown on an ABA supplemented medium (Supplemental Fig. S11B). Taken together with our pharmacological data, these datasets indicate AtSLP2 functions to negatively regulate GA-related cellular processes.
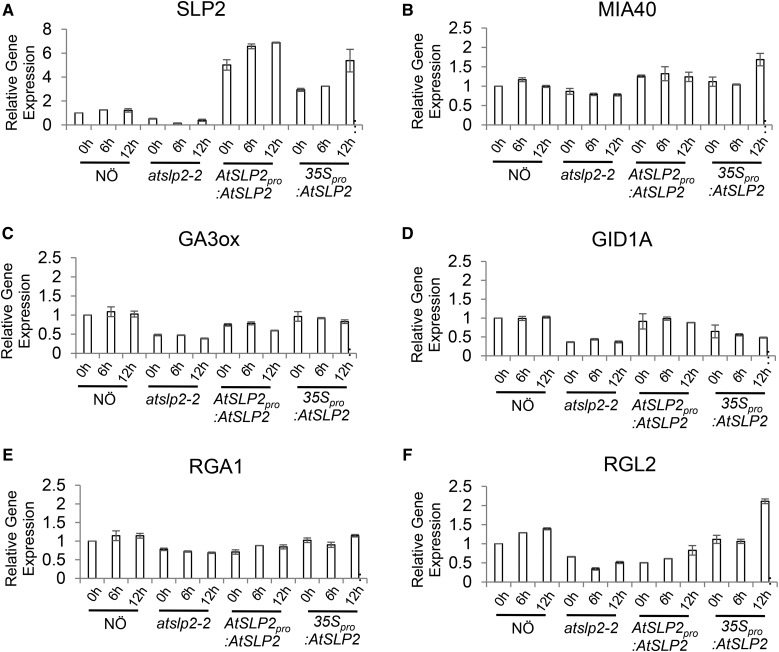
Subsequent assessment of Arabidopsis MIA40, GA3OX, GID1, RGA1, and RGL2 transcript levels in atslp2-2, AtSLP2pro::AtSLP2, wild-type NÖ and 35Spro::AtSLP2 seeds over a 12 h imbibition time course by qRT-PCR (Fig. 6) further corroborated available microarray data of germinating Arabidopsis seeds (Supplemental Figs. S10 and S11). Consistent with the presence, absence, or overabundance of AtSLP2 protein, AtSLP2 transcript levels changed as expected (Fig. 6A). AtMIA40 transcript levels largely mirrored these transcriptional changes in AtSLP2, with moderately decreased expression in atslp2-2 and slightly increased expression in 35Spro::AtSLP2 (Fig. 6B). GA biosynthetic and signaling machinery transcript levels similarly exhibited changes in transcript levels mirroring AtSLP2 (Fig. 6, C–F). This was most well resolved in atslp2-2, where decreases in both GA biosynthetic and signaling machinery were observed. The largest changes in transcript levels were observed for AtSLP2 in the 35Spro::AtSLP2 and AtSLP2pro::AtSLP2 plant lines (4–5 RQ fold increase), while overall changes in GA-related transcript levels were relatively moderate, exhibiting only a ±0.5 to 1.0 RQ fold-change. Changes in AtSLP2 transcript abundance were paralleled by a corresponding change in AtSLP2 protein levels (Supplemental Fig. S8), while peak AtSLP2 and AtMIA40 transcript levels (∼6 h imbibition) corresponded to peak AtSLP2 protein (3–6 h) during wild-type NÖ imbibition (Fig. 6; Supplemental Fig. S8).
Figure 6.
Quantitative PCR analysis of GA biosynthetic and signaling protein genes from imbibed atslp2-2, wild-type NÖ, AtSLP2pro::AtSLP2, and 35Spro::AtSLP2 seeds. Seeds were imbibed for 0 h, 6 h, and 12 h in water at 4°C in the dark followed by snap freezing. A to F, Transcriptional expression of Arabidopsis SLP2, MIA40, GA3 oxidase (GA3OX), GA-insensitive dwarf 1a (GID1A), regulator of GA1 (RGA1), and regulator of GA-like 2 (RGL2) was assessed. Error bars represent ± se (n = 3).
DISCUSSION
AtSLP2 Phosphatase Represents a Novel IMS Client of Redox Relay Protein AtMIA40
SLP2 represents a novel interaction partner of MIA40 in photosynthetic eukaryotes, as SLP2 phosphatases are absent in metazoans (Uhrig and Moorhead, 2011). Previous examination of atmia40 insertional mutant plants found AtMIA40 responsible for importing the chaperone for Mn-superoxide dismutase (CCS1) and Cu/Zn superoxide dismutase 1 into the mitochondrial IMS, as has been observed in other eukaryotes (Kawamata and Manfredi, 2010), as well as Cu/Zn superoxide dismutase 3 into the plant peroxisome (Hentchel and Escalante-Semerena, 2015). In addition to these proteins, research in nonphotosynthetic eukaryotes has identified COX17, translocase of inner membrane proteins TIM22, and a number of other mitochondrial IMS proteins to be directly regulated by MIA40 (Banci et al., 2009; Reddehase et al., 2009; Sideris et al., 2009; Darshi et al., 2012; Weckbecker et al., 2012). However, AtSLP2 represents the first protein phosphatase identified to interact with MIA40.
MIA40 proteins have been found to catalyze the formation of disulfide bridges on target protein substrates through their oxidoreductase activity, trapping the MIA40 substrate in the mitochondrial IMS (Banci et al., 2009; Sideris et al., 2009). This is achieved via the key cysteines of MIA40’s active site (the CPC motif), which first forms disulfide bridge intermediates with the target protein substrate, followed by the formation of disulfide bridges on the target substrate and the release of reduced MIA40. The association of AtSLP2 with AtMIA40 appears to be disulfide bond dependent as thiol modification of reduced cysteines blocked their association (Fig. 3A). Through sequence comparison with its closely related paralog SLP1, we deduced and then confirmed which Cys pairs form disulfide bonds and are necessary for AtMIA40 activation of AtSLP2 (Fig. 3, C and D). Collectively, these findings support AtMIA40 activation of AtSLP2 through the catalyzed formation of disulfide bridges. Like other MIA40 substrates, this may function to trap AtSLP2 in the IMS by providing correct tertiary structure (Fischer and Riemer, 2013).
The mitochondrial localization of AtSLP2 (Fig. 2), combined with its 4- (pNPP) to 20- (phosphorylated peptides) fold increase in activity upon interaction with IMS-targeted AtMIA40 and a phosphorylated substrate preference indicative of Ser/Thr PPP-family protein phosphatases (Fig. 4) suggest reversible protein phosphorylation plays a regulatory role within the mitochondrial IMS. No studies to date have specifically examined the role of reversible protein phosphorylation in the mitochondrial IMS of any organism despite targeted proteomics studies revealing the presence of a number of protein kinases and protein phosphatases in this subcompartment (Salvi et al., 2005; Pagliarini and Dixon, 2006; Vögtle et al., 2012; Duncan et al., 2013). To date, 10 protein kinases have been localized to the plant mitchondrion, but none specifically to the IMS (Havelund et al., 2013). Furthermore, recent elucidation of the IMS proteome of yeast identified a number of IMS proteins that have been previously documented in whole mitochondria phosphoproteomics studies to be phosphorylated (Vögtle et al., 2012). Given the prevalence of regulatory protein phosphorylation across eukaryotes, it is reasonable to predict that mitochondrial IMS reversible protein phosphorylation also represents a conserved phenomenon.
Negative Regulation of Seed Germination through GA Metabolism
In exploring the biological function of AtSLP2, atslp2-2 seeds exhibited accelerated seed germination compared to NÖ seeds, while 35Spro::AtSLP2 seeds displayed delayed seed germination (Fig. 5). This indicated that AtSLP2 was a regulator of seed germination. With germinative processes predominantly governed by ABA and GA, this suggested that atslp2-2 seeds were either ABA insensitive/underproducing or GA sensitive/overproducing. Lack of dramatic ABA insensitivity and maintenance of atslp2-2-accelerated germination in the presence of ABA indicated that the observed seed germination phenotype is likely due to enhanced GA production or altered GA signaling. If atslp2-2 seeds were ABA insensitive, application of 1 µm ABA should not have induced the concentration-dependent effect on seed germination that was documented previously (Koornneef et al., 1989). Furthermore, Uniconazole almost completely abrogated the accelerated germination of atslp2-2 seeds, indicating GA is required for the observed germination phenotypes. Given GA signaling (negative regulators) mutants such as rgl2 and other DELLA loss-of-function mutants can germinate in the presence of GA inhibitors and confer GA phenotypes in the absence of bioactive GA (Sun and Kamiya, 1994; Lee et al., 2002; Cao et al., 2005), the AtSLP2-AtMIA40 complex likely functions to negatively regulate GA biosynthesis.
If GA biosynthesis were up-regulated in atslp2-2 seeds, we would expect a corresponding increase in GA-induced degradation of negative regulators RGL2 and RGA1 (Xu et al., 2014). In accordance with this hypothesis, expression of DELLA protein RGL2, the major determinant of seed germination, was down-regulated in atslp2-2 (Fig. 6; Lee et al., 2002). In 35Spro::AtSLP2 seeds, this pattern of GA-negative regulator down-regulation was inverse. 35Spro::AtSLP2 seeds maintained elevated RGL2 transcript levels relative to wild-type Arabidopsis following imbibition (Fig. 6).
AtSLP2-AtMIA40 Protein Complex Functions to Negatively Regulate GA Biosynthesis
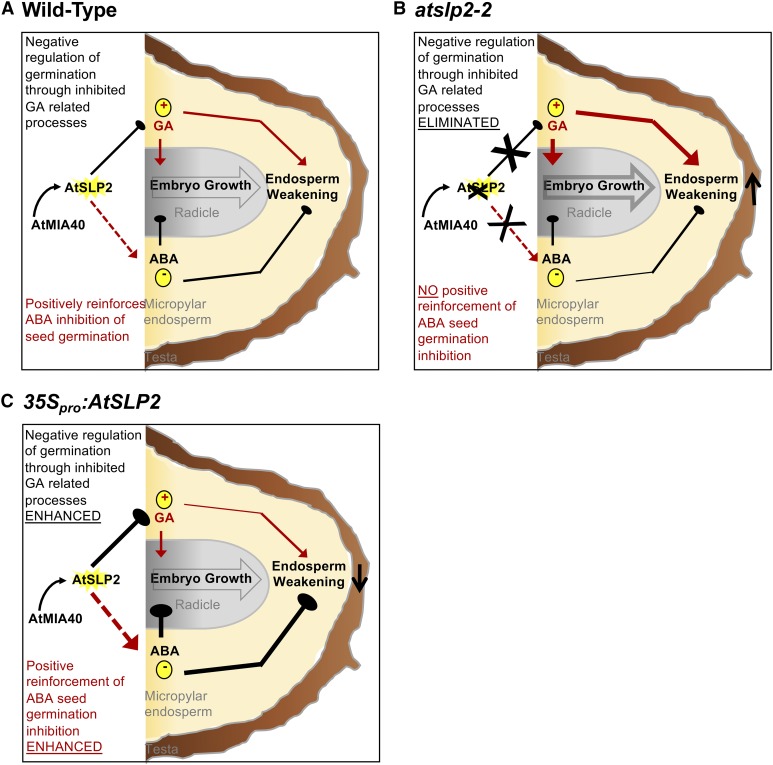
When compared to atslp2-2 seeds, atmia40 seeds exhibited a moderate accelerated germination phenotype (Supplemental Fig. S12). This suggests that AtSLP2 likely represents the functional driver of the AtSLP2-AtMIA40 complex in negatively regulating GA-related cellular processes (Fig. 7). Reinforcing this hypothesis is the bacterially expressed and purified AtSLP2, which maintained detectable protein phosphatase activity even in the absence of AtMIA40 (Uhrig and Moorhead, 2011), indicating that atmia40 seeds likely possess some residual AtSLP2 protein phosphatase activity, which mitigates, to a certain extent, the absence of AtMIA40 and its requirement to fully activate AtSLP2. With seed phenotype data supporting a role for mitochondrially targeted AtSLP2 in regulating cytosolic GA-related processes, there remains a need to resolve the intermediate mechanisms connecting the mitochondria to cytosolic GA biosynthesis.
Figure 7.
Model of AtSLP2-AtMIA40 protein complex function during seed germination. A, Under wild-type conditions, AtMIA40-activated AtSLP2 functions to negatively regulate GA biosynthesis and in doing so positively reinforces ABA inhibition of seed germination. B, Absence of AtSLP2 (atslp2-2) results in unimpeded GA-related cellular processes leading to an accelerated seed germination phenotype. C, Overexpression of AtSLP2 (35Spro::AtSLP2) leads to enhanced inhibition of GA-related cellular processes, which concomitantly enhances ABA inhibition of seed germination. This leads to a delayed seed germination phenotype. Effects of the presence or absence of an AtSLP2-AtMIA40 protein complex over seed germinative processes is described by a combination of arrow width and color. Arrow width denotes enhanced (thick) versus reduced (thin) pathway flux/phytohormone effect, respectively. Solid black lines versus solid red lines depict negative or positive influence over seed germination, respectively. Dashed red lines represent indirect AtSLP2-AtMIA40 protein complex influence over ABA-directed seed germination processes.
With the discovery that AtSLP2 protein phosphatase interacts with IMS redox relay protein AtMIA40 to negatively regulate seed germination via GA-related processes, there exists a pressing need to identify its substrate(s). Albeit challenging, substrate identification will better resolve the working hypothesis presented here by shedding light on the connection between mitochondrial targeted AtSLP2 and its negative regulation of cytosolic/nuclear-localized GA-related processes. The work presented here outlines an exciting new connection between the mitochondria and cytosol through plant cell signaling, in addition to providing precedence for reversible protein phosphorylation in the mitochondrial IMS. Future studies will focus on elucidating the intermediate steps and components of how AtSLP2 negatively regulates GA-related processes.
MATERIALS AND METHODS
Plant Growth Conditions
Standard Arabidopsis (Arabidopsis thaliana) growth conditions involved dry sterilization and stratification at 4°C for 2 d in the dark on 0.5× Murashige and Skoog (MS) agar plates. Plants were germinated under 16 h light and 8 h dark at 22°C at a light level of 125 µmol m−2s−1. Five-day-old seedlings were transferred to soil supplemented with 0.5 g/L 20:20:20 fertilizer and grown under a 12-h-light/12-h-dark cycle regiment at 22°C at a light level of 125 µmol m−2s−1. Plant roots were isolated from germinating Arabidopsis seedlings grown as above in Magenta boxes for 21 d. Magenta box growth media consisted of 0.5× MS media supplemented with 1 mm potassium phosphate. Seedlings subjected to ABA and Uniconazole (LKT Laboratories) were sterilized and stratified as above on 0.5× MS agar followed by germination under 16 h light and 8 h dark at 22°C for 4 d. For seed germination assays, statistical significance between mutant and corresponding wild-type seed germination was performed using a two-tailed Student’s t test. Sample variance was assessed using an F-test. No statistically significant variance was observed.
Insertional atslp2 and atmia40 Plant Genotyping
Insertional mutants atslp2-2 (RATM13-2055-1) and atmia40 (SALK_044358) Arabidopsis were screened by PCR using the following gene primers: 5′-TGGTTGCAATTCTGACACAAC, 3′-CAAGAATCTCTTTGCAATCGG (atslp2-2), and 5′-TCGTACAGATGCGATCATCTG, 3′-AAGGGACACAAAACGTAAACG (atmia40). The T-DNA insert primers used were: 3′-GTATTTATCCCGTTCGTTTTCGT (atslp2-2) and 5′-TGGTTCACGTAGTGGGCCATCG (atmia40). Insertional mutant plant lines were obtained from the Riken Bioresource Center Experimental Plant Division (RIKEN; http://epd.brc.riken.jp/en/) and the Arabidopsis Information Resource (www.arabidopsis.org; atmia40), respectively.
Molecular Cloning, Expression, and Recombinant Protein Purification
Fluorescent and TAP constructs were created using full-length At1g18480 (AtSLP2), At1g07010 (AtSLP1), and At5g23395 (AtMIA40) clones obtained from the Arabidopsis Information Resource (http://www.arabidopsis.org/). PCR amplification of each respective cDNA was performed using Gateway-compatible primers and inserted into pDONR221 (Invitrogen). Fluorescent constructs were subsequently created using the binary vectors pB7RWG2 (cRFP) and pK7RWG2 (cGFP; https://gateway.psb.ugent.be/), while TAP clones were created using pYL436 (Rubio et al., 2005). Transformation of each construct into Agrobacterium tumefaciens strain GV3101 (A. tumefaciens) was performed to facilitate stable integration of each construct into either Arabidopsis plants (Zhang et al., 2006) or cell culture (Templeton et al., 2011).
At5g23395 was cloned into pDEST15 (N-terminal GST-tag) and pDEST17 (N-terminal HIS6-tag) for expression in and purification from Escherichia coli, while AtSLP1 and 2 were cloned into pET-48b(+) (Novagen) and purified via Ni-NTA as previously described (Uhrig and Moorhead, 2011). GST-only (pGEX4T-1; Qiagen) was also expressed and purified for use as a control in enzyme and in vitro interaction analyses. GST-AtMIA40 and GST-only expression utilized BL21 (DE3) Codon(+)-RIL (Stratagene) E. coli grown at 37°C to an OD600 of 0.4 to 0.5 followed by induction with 0.1 mm isopropyl β-d-1-thiogalactopyranoside overnight (18 h) at room temperature (23°C). Protein purification for use in enzymatic assays and protein interaction experimentation was performed as per Uhrig and Moorhead (2011) using 500 µL of settled glutathione Sepharose (GE Healthcare). Protein elution was performed using 10 mm GSH in 50 mm HEPES-KOH, pH 7.8, 150 mm NaCl and 5% glycerol. Eluates were concentrated to a small volume using a 10,000 D cutoff concentrator (Amicon), followed by dialysis against elution buffer lacking GSH prior to snap freezing and storage at −80°C.
Complementation and Screening of AtSLP2 Mutant Plants
Complementation of atslp2-2 knockout plants was performed by the amplification of AtSLP2 native promoter and gene from genomic DNA of wild-type ecotype NÖ Arabidopsis leaves using the Gateway-compatible primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCTATC TCTCTTATAACGAAGAAA-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCT GGGTCAGCTTTGACTTCAACTTGCTTAG-3′. The 1.581-kb promoter region was determined based on the Arabidopsis Gene Regulatory Information Server. The fragment was inserted into pDONR221 and cloned into the destination vector pEarleyGate 304 (Earley et al., 2006). The binary vector was transformed into atslp2-2 mutant as above. Basta (BaR)-resistant plants were genotyped by PCR and expression of AtSLP2 assessed by western blotting.
Arabidopsis Cell Culture Transfection and TAP Pull-Downs
Wild-type light-grown Arabidopsis cell culture was individually transfected by A. tumefaciens carrying AtSLP2, AtSLP1, AtMIA40, and GFP-TAP constructs as previously described (Uhrig and Moorhead, 2011). Positive transformants were screened via immunoblotting with anti-Myc IgG (ICL). Arabidopsis cell culture was subsequently transitioned from light to dark over a 2 week period, with subculturing performed every 7 d. TAP pull-downs from dark cell culture were performed as previously described (Templeton et al., 2011) using an extract from 50 g of cells per pull-down, while pull-downs from roots were performed using 5 g of tissue. Subsequent washing of tagged protein bound Ni-NTA with 10 mL of triethylammonium bicarbonate (pH 8.5) was performed prior to on-bead, overnight (18 h) proteolysis using 750 ng of trypsin (Promega) at 30°C. Peptides were eluted using a 50% acetonitrile: 2.5% formic acid solution and dried prior to analysis by mass spectrometry. TAP pull-downs subjected to immunoblot analysis were eluted from Ni-NTA using a minimal volume of 1× sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and boiling for 5 min.
Mass Spectrometry
Sample digestion, processing, mass spectrometry, and data analysis were performed as in Templeton et al. (2011). In addition to GFP-TAP pull-downs controlling for experimentation involving cell culture, AtSLP1 offered a control for AtSLP2 pull-downs from root tissue, as AtSLP1 is not expressed in nongreen tissues and is the closest ortholog of AtSLP2 (Uhrig and Moorhead, 2011). Bona fide binding partners were only considered if they were not found in the majority of independent negative controls: 6× GFP-TAP and 7× AtSLP1-TAP pull-downs. Raw data from the excised band and on-bead digested samples have been deposited at “MassIVE” (massive.ucsd.edu).
AtMIA40 Immunoprecipitation
Affinity-purified anti-AtMIA40 IgG and rabbit preimmune serum IgG (10 µg) were coupled to Protein A-Sepharose 4B beads (Life Technologies) as described in Ulke-Lemée et al. (2007). AtMIA40 immunoprecipitation was performed with 50 mg protein extract isolated from 10-d-old dark-grown Arabidopsis cell culture as in Uhrig et al. (2016). Protein extract was pretreated with 20 mm DTT for 45 min at 4°C followed by alkylation with 110 mm NEM for 1 h at 4°C. A control without reducing/alkylating agents was performed in parallel. Immunoprecipitations were performed three times with three separate plant samples.
Transient Vicia faba Expression and Imaging
Transient expression of 35Spro::AtSLP2-RFP and 35Spro::Peroxisome-GFP or 35Spro::Mito-GFP (Nelson et al., 2007) in V. faba leaves was achieved by particle bombardment (PDS-1000 system; Bio-Rad). Plasmid DNA (5 µg each for cobombardment or 10 µg for single bombardment) was coupled to 1 mg of gold microcarriers, washed, spotted on a macrocarrier, and accelerated onto the leaves. Stably transfected plants expressing 35Spro::AtSLP2-GFP were stained with 10 µm MitoTracker Deep Red FM for 10 min and rinsed with water. All stably transfected 35Spro::AtMIA40-RFP/35Spro::AtSLP2-GFP Arabidopsis tissues were mounted on a microscope slide in 50% (v/v) glycerol and visualized at 63× magnification using a confocal microscope (Leica TCS SP5). The following excitation/emission (Ex/Em) settings were used: RFP, 543/580 to 682 nm; GFP, 488/497 to 538 nm; MitoTracker Deep Red FM, 633/700 to 756 nm.
Isolation of Arabidopsis Cell Culture Mitochondrial Fractions and Peroxisomes
Seven-day-old, dark-grown Arabidopsis culture (120 g) was filtered through one layer of Miracloth to remove growth medium and homogenized batchwise 40 g at a time in a precooled 4°C mortar and pestle. Grinding of each 40 g was performed in 100 mL homogenization buffer consisting of 0.3 m mannitol, 50 mm MOPS-KOH, pH 7.8, 5 mm EDTA, 0.5% (w/v) BSA, 1.0% (w/v) polyvinlypyrrolidone, and 10 mm DTT added just before use. Isolation of mitochondria plus peroxisome was obtained by Percoll density gradient centrifugation as previously described (Eubel et al., 2007). Separation of mitochondria from peroxisome was achieved by resuspending the fraction in 1 mL Suc buffer (0.3 m Suc, 10 mm TES-NaOH, pH 7.5, 0.1% [w/v] BSA) and overlaying on 28% (v/v) Percoll in Suc buffer followed by centrifugation at 41,000g for 30 min at 4°C (Struglics et al., 1993). Mitochondria (upper layer) and peroxisome (lower layer) fractions were collected separately, washed in mannitol buffer (20 mm MOPS-KOH, pH 7.5, 0.3 m mannitol, 0.2% [w/v] BSA) and centrifuged at 18,675g for 15 min at 4°C. Isolated mitochondria and peroxisomes were lysed with 50 mm MOPS-KOH, pH 7.5, 150 mm NaCl, 0.2% (v/v) Triton X-100 and boiled in SDS-cocktail. Mitochondrial subcompartments were isolated as in Sweetlove et al. (2001). Organelle isolations were performed three times from three separate plant samples and a representative experiment is shown.
Enzymatic Analysis
Enzyme assays utilizing pNPP (Sigma-Aldrich) were conducted using Ni-NTA-purified HIS6-AtSLP2 and glutathione Sepharose-purified GST-AtMIA40 or HIS6-AtMIA40 (Supplemental Fig. S2). All assays were done in parallel with Ni-NTA-purified eluates generated from nontransformed BL21 (DE3) Codon(+)-RIL E. coli to account for potential background pNPP cleavage from nonspecific copurifying proteins (Supplemental Fig. S2). Phosphatase activity was calculated relative to assays conducted with AtSLP2 only under previously determined conditions giving highest activity (Uhrig and Moorhead, 2011). All pNPP assays were conducted as previously described (Uhrig and Moorhead, 2011) using either 5 mm DTT, Tris(2-carboxyethyl)phosphine or reduced glutathione as a reductant. Expressed and purified GST only was used as an assay control.
Malachite green (Sigma-Aldrich) assays were performed as previously described (Baykov et al., 1988), alternatively employing a 10% (w/v) ammonium molybdate solution. Each assay used 1 µg of recombinant phosphatase protein incubated for 1 h at 30°C in 160 µL of 1× dilution buffer containing each respective experimental phosphopeptide substrate (Supplemental Table S2). Quenched assays were left at room temperature (23°C) for 10 min prior to spectrophotometric assessment at 630 nm. Several phosphorylated peptide substrates were the kind gift of Dr. D. Alessi of the Protein Phosphorylation Unit (Dundee, Scotland).
Assays performed using TAP isolated AtSLP2 employed 65 µL of protein phosphatase bound to Ni-NTA that was isolated from 20 g of Arabidopsis rosette tissue as outlined above. Each aliquot of coupled Ni-NTA matrix was resuspended in 235 µL of 1× dilution buffer containing 500 µm of the respective phosphorylated peptide and 1 µm HIS6-AtMIA40. Each assay was incubated for 1 h in a 30°C water bath, followed by pelleting of Ni-NTA matrix. Reaction mix was removed (160 µL) and quenched with 40 µL of malachite green solution.
Anti-AtMIA40 Antibody Production
HIS6-AtMIA40 was expressed in BL21 (DE3) Codon(+)-RIL E. coli at 37°C, induced with 0.5 mm IPTG for 4 h and purified from inclusion bodies using Ni-NTA according to the manufacturer’s instructions (Qiagen). Purified protein was dialyzed against water and used for polyclonal antibody production in a New Zealand White rabbit as described (Tran et al., 2004). AtMIA40 antigen purity and crude immune serum specificity are shown (Supplemental Figs. S3 and S4).
Seed and Root Protein Extraction
Seeds (50 mg) were extracted by mortar and pestle in a solution consisting of 100 mm Tris-HCl, pH 9.5, 150 mm NaCl, 1% (w/v) sarkosyl, and 5 mm DTT. Root proteins from 10-d-old seedlings were extracted with 50 mm HEPES-NaOH, pH 7.5, 150 mm NaCl, 5% (v/v) glycerol, 1% (v/v) Tween 20, 1 mm PMSF, and 1 mm benzamidine. Seed and root lysates were clarified by centrifugation at 4°C and 14,000g.
Western Blotting
All antibodies were validated within this manuscript (MIA40) or as noted below in published articles or in the datasheets for the commercial antibodies. Affinity-purified polyclonal rabbit anti-AtSLP1 and 2 antibodies previously generated (Uhrig and Moorhead, 2011) were used at 1.5 and 1.2 µg/mL, respectively. Rabbit anti-AtMIA40 crude immune serum was used at a 1:1000 dilution and validated in Supplemental Figs. S3 and S4. Mitochondria and peroxisomes were probed using 1:100 antimitochondrial pyruvate dehydrogenase complex E1α (mitochondrial matrix [Szurmak et al., 2003]) and 1:200 anticatalase (peroxisome) IgG (Chuong et al., 2005). Mitochondrial subcompartments were blotted using 1:5000 anti-H protein of Gly decarboxylase complex (anti-GDC-H; mitochondria matrix marker; Agrisera/AS05 074), 1:5000 antivoltage-dependent anion-selective channel protein 1 (anti-VDAC1; OMM marker; Agrisera) and 2 µg/mL anticytochrome c (anticyt c [Sweetlove et al., 2001]) IgG. Commercially available rabbit anti-Myc (ICL/RMYC-45A) and mouse anti-GFP (Roche/11814460001) antibodies were used at 0.2 µg/mL and 0.4 µg/mL, respectively.
qRT-PCR Analysis
Total RNA from the various Arabidopsis seed lines were isolated using a modified TRIzol (Invitrogen) protocol (Meng and Feldman, 2010). First-strand cDNA was synthesized from 1.67 μg total RNA using oligo(dT)12-18 primer and SuperScript II Reverse Transcriptase (Invitrogen) following the manufacturer’s instructions. The qPCR was performed using StepOnePlus Real-Time PCR System (Applied Biosystems). Primer pairs are listed (Supplemental Table S3). Each PCR reaction contained 1× Fast SYBR Green Master Mix (Applied Biosystems), 200 nm of each primer, and 0.25 μL cDNA in a final volume of 20 μL. PCR amplification was performed for 40 cycles at 95°C for 3 s and 60°C for 30 s with a preceding initial enzyme activation of 20 s at 95°C. Relative expression levels were calculated by Δ-ΔCt method, and all quantifications were normalized using At1g13440 mRNA as an internal control (Dekkers et al., 2012). For each target gene, the reactions were carried out in triplicate.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At5g23395, At1g18480, At1g07010, At5g25900, At3g05120, At1g14920, At3g03450, AT3G13860, AT1G72730, and AT5G37600.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Immunoblot analysis of dark-grown, wild-type Arabidopsis cell culture.
Supplemental Figure S2. Colloidal blue-stained purified fractions of bacterially expressed and purified Arabidopsis proteins used during in vitro experimentation.
Supplemental Figure S3. Purification of 6 m urea extracted HIS6-AtMIA40.
Supplemental Figure S4. Analysis of anti-AtMIA40 crude immune serum.
Supplemental Figure S5. Confocal imaging of AtSLP2 and AtMIA40 in plant cells.
Supplemental Figure S6. SDS-PAGE analysis of purified HIS6-AtSLP2 and AtMIA40.
Supplemental Figure S7. Immunoblot analysis of atslp2-2, wild-type NÖ, AtSLP2pro:AtSLP2 complemented atslp2-2 and 35Spro:AtSLP2 Arabidopsis seed and root tissue.
Supplemental Figure S8. Western blot analysis of AtSLP2 and AtMIA40 expression in imbibed and germinating seeds.
Supplemental Figure S9. Quantitative time-course measurement of seed germination on 0.5× MS-agar plates containing ABA or Uniconazole.
Supplemental Figure S10. Relative transcriptional expression of GA-related biosynthetic and signaling proteins from Biological Arabidopsis Resource.
Supplemental Figure S11. Relative transcriptional changes in GA-related biosynthetic and signaling proteins as well as AtSLP2 from Genevestigator.
Supplemental Figure S12. Quantitative measurement of seed germination on 0.5× MS-agar plates containing ABA or Uniconazole.
Supplemental Table S1. AtSLP2-specific protein interactors recovered from cell culture and root tissue.
Supplemental Table S2. Phosphorylated peptide substrates used in AtSLP2 assays.
Supplemental Table S3. Primers for qPCR analysis of GA-related genes.
Supplemental Table S4. SLP1 and SLP2 sequence data used for MAFFT alignment.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Douglas Randall (University of Missouri) for his kind provision of mitochondrial PDC E1α antibodies. Thanks also go to Dr. D. Alessi of the Protein Phosphorylation Unit (Dundee, Scotland) for the provision of phosphorylated peptides and Siyu Liang for helpful discussions regarding qRT-PCR. Finally, thanks to Dr. Doug Muench and Dr. Howard Ceri (University of Calgary) for providing fluorescent DNA constructs and confocal microscope facilities.
Glossary
- TAP
tandem affinity purification
Footnotes
This work was funded by the National Science and Engineering Research Council of Canada, Alberta Ingenuity Technology Futures, and the Killam Trusts.
Articles can be viewed without a subscription.
References
- Andreeva AV, Kutuzov MA (2004) Widespread presence of “bacterial-like” PPP phosphatases in eukaryotes. BMC Evol Biol 4: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cefaro C, Ciofi-Baffoni S, Gallo A, Martinelli M, Sideris DP, Katrakili N, Tokatlidis K (2009) MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat Struct Mol Biol 16: 198–206 [DOI] [PubMed] [Google Scholar]
- Baykov AA, Evtushenko OA, Avaeva SM (1988) A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem 171: 266–270 [DOI] [PubMed] [Google Scholar]
- Cao D, Hussain A, Cheng H, Peng J (2005) Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223: 105–113 [DOI] [PubMed] [Google Scholar]
- Chuong SD, Park NI, Freeman MC, Mullen RT, Muench DG (2005) The peroxisomal multifunctional protein interacts with cortical microtubules in plant cells. BMC Cell Biol 6: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darshi M, Trinh KN, Murphy AN, Taylor SS (2012) Targeting and import mechanism of coiled-coil helix coiled-coil helix domain-containing protein 3 (ChChd3) into the mitochondrial intermembrane space. J Biol Chem 287: 39480–39491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJ, Willems L, Bassel GW, van Bolderen-Veldkamp RP, Ligterink W, Hilhorst HW, Bentsink L (2012) Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53: 28–37 [DOI] [PubMed] [Google Scholar]
- Duncan MR, Fullerton M, Chaudhuri M (2013) Tim50 in Trypanosoma brucei possesses a dual specificity phosphatase activity and is critical for mitochondrial protein import. J Biol Chem 288: 3184–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Eubel H, Heazlewood JL, Millar AH (2007) Isolation and subfractionation of plant mitochondria for proteomic analysis. Methods Mol Biol 355: 49–62 [DOI] [PubMed] [Google Scholar]
- Fischer M, Riemer J (2013) The mitochondrial disulfide relay system: roles in oxidative protein folding and beyond. Int J Cell Biol 2013: 742923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelund JF, Thelen JJ, Møller IM (2013) Biochemistry, proteomics, and phosphoproteomics of plant mitochondria from non-photosynthetic cells. Front Plant Sci 4: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K. (2008) The Erv1-Mia40 disulfide relay system in the intermembrane space of mitochondria. Biochim Biophys Acta 1783: 601–609 [DOI] [PubMed] [Google Scholar]
- Hentchel KL, Escalante-Semerena JC (2015) Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol Mol Biol Rev 79: 321–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heroes E, Lesage B, Görnemann J, Beullens M, Van Meervelt L, Bollen M (2013) The PP1 binding code: a molecular-lego strategy that governs specificity. FEBS J 280: 584–595 [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Riemer J (2012) Mitochondrial disulfide relay: redox-regulated protein import into the intermembrane space. J Biol Chem 287: 4426–4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJ (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Kawamata H, Manfredi G (2010) Import, maturation, and function of SOD1 and its copper chaperone CCS in the mitochondrial intermembrane space. Antioxid Redox Signal 13: 1375–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Hilhorst HW, Karssen CM (1989) In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol 90: 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera B, Cohn MA, Leubner-Metzger G (2005) Plant hormone interactions during seed dormancy release and germination. Seed Sci Res 15: 281–307 [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Feldman L (2010) A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds. Biotechnol J 5: 183–186 [DOI] [PubMed] [Google Scholar]
- Moorhead GB, De Wever V, Templeton G, Kerk D (2009) Evolution of protein phosphatases in plants and animals. Biochem J 417: 401–409 [DOI] [PubMed] [Google Scholar]
- Mordas A, Tokatlidis K (2015) The MIA pathway: a key regulator of mitochondrial oxidative protein folding and biogenesis. Acc Chem Res 48: 2191–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Tintelnot S, Leubner-Metzger G (2006) Endosperm-limited Brassicaceae seed germination: Abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol 47: 864–877 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Dixon JE (2006) Mitochondrial modulation: Reversible phosphorylation takes center stage? Trends Biochem Sci 31: 26–34 [DOI] [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L (2008) The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddehase S, Grumbt B, Neupert W, Hell K (2009) The disulfide relay system of mitochondria is required for the biogenesis of mitochondrial Ccs1 and Sod1. J Mol Biol 385: 331–338 [DOI] [PubMed] [Google Scholar]
- Rubio V, Shen Y, Saijo Y, Liu Y, Gusmaroli G, Dinesh-Kumar SP, Deng XW (2005) An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J 41: 767–778 [DOI] [PubMed] [Google Scholar]
- Salvi M, Brunati AM, Toninello A (2005) Tyrosine phosphorylation in mitochondria: a new frontier in mitochondrial signaling. Free Radic Biol Med 38: 1267–1277 [DOI] [PubMed] [Google Scholar]
- Sharma K, D’Souza RC, Tyanova S, Schaab C, Wiśniewski JR, Cox J, Mann M (2014) Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Reports 8: 1583–1594 [DOI] [PubMed] [Google Scholar]
- Sideris DP, Petrakis N, Katrakili N, Mikropoulou D, Gallo A, Ciofi-Baffoni S, Banci L, Bertini I, Tokatlidis K (2009) A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J Cell Biol 187: 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struglics A, Fredlund KM, Rasmusson AG, Moller IM (1993) The presence of a short redox chain in the membrane of intact potato tuber peroxisomes and the association of malate dehydrogenase with the peroxisomal membrane. Physiol Plant 88: 19–28 [Google Scholar]
- Sun TP, Kamiya Y (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Mowday B, Hebestreit HF, Leaver CJ, Millar AH (2001) Nucleoside diphosphate kinase III is localized to the inter-membrane space in plant mitochondria. FEBS Lett 508: 272–276 [DOI] [PubMed] [Google Scholar]
- Szurmak B, Strokovskaya L, Mooney BP, Randall DD, Miernyk JA (2003) Expression and assembly of Arabidopsis thaliana pyruvate dehydrogenase in insect cell cytoplasm. Protein Expr Purif 28: 357–361 [DOI] [PubMed] [Google Scholar]
- Templeton GW, Nimick M, Morrice N, Campbell D, Goudreault M, Gingras AC, Takemiya A, Shimazaki K, Moorhead GB (2011) Identification and characterization of AtI-2, an Arabidopsis homologue of an ancient protein phosphatase 1 (PP1) regulatory subunit. Biochem J 435: 73–83 [DOI] [PubMed] [Google Scholar]
- Tran HT, Ulke A, Morrice N, Johannes CJ, Moorhead GB (2004) Proteomic characterization of protein phosphatase complexes of the mammalian nucleus. Mol Cell Proteomics 3: 257–265 [DOI] [PubMed] [Google Scholar]
- Uhrig RG, Kerk D, Moorhead GB (2013a) Evolution of bacterial-like phosphoprotein phosphatases in photosynthetic eukaryotes features ancestral mitochondrial or archaeal origin and possible lateral gene transfer. Plant Physiol 163: 1829–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig RG, Labandera AM, Moorhead GB (2013b) Arabidopsis PPP family of serine/threonine protein phosphatases: many targets but few engines. Trends Plant Sci 18: 505–513 [DOI] [PubMed] [Google Scholar]
- Uhrig RG, Labandera AM, Muhammad J, Samuel M, Moorhead GB (2016) Rhizobiales-like phosphatase 2 from Arabidopsis thaliana is a novel phospho-tyrosine-specific phospho-protein phosphatase (PPP) family protein phosphatase. J Biol Chem 291: 5926–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig RG, Moorhead GB (2011) Two ancient bacterial-like PPP family phosphatases from Arabidopsis are highly conserved plant proteins that possess unique properties. Plant Physiol 157: 1778–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulke-Lemée A, Trinkle-Mulcahy L, Chaulk S, Bernstein NK, Morrice N, Glover M, Lamond AI, Moorhead GB (2007) The nuclear PP1 interacting protein ZAP3 (ZAP) is a putative nucleoside kinase that complexes with SAM68, CIA, NF110/45, and HNRNP-G. Biochim Biophys Acta 1774: 1339–1350 [DOI] [PubMed] [Google Scholar]
- Vögtle FN, Burkhart JM, Rao S, Gerbeth C, Hinrichs J, Martinou JC, Chacinska A, Sickmann A, Zahedi RP, Meisinger C (2012) Intermembrane space proteome of yeast mitochondria. Mol Cell Proteomics 11: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckbecker D, Longen S, Riemer J, Herrmann JM (2012) Atp23 biogenesis reveals a chaperone-like folding activity of Mia40 in the IMS of mitochondria. EMBO J 31: 4348–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Liu Q, Yao T, Fu X (2014) Shedding light on integrative GA signaling. Curr Opin Plant Biol 21: 89–95 [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.