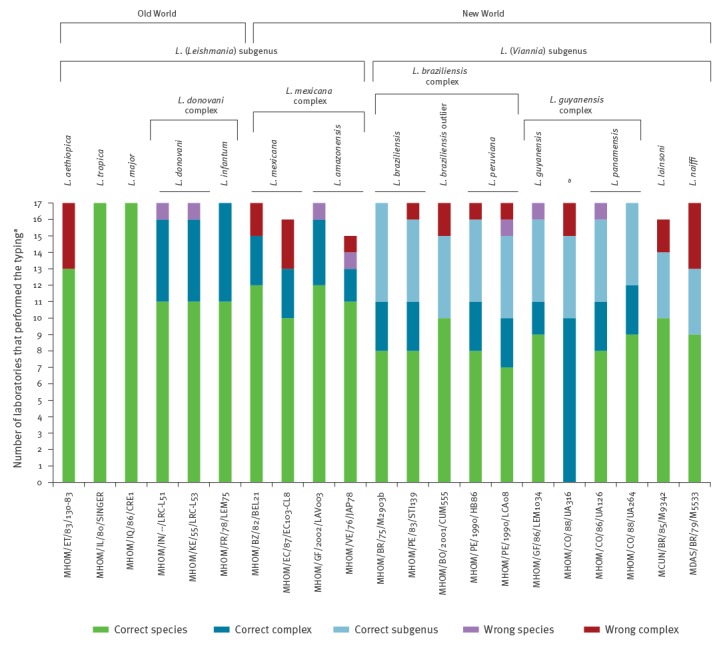
Figure 2.
Typing results for each of the 21 strains included in study comparing Leishmania typing results in 16 European clinical laboratories, 2014
MLSA: multilocus sequence analysis; WHO: World Health Organization.
a One laboratory reported the use of two separate methods.
b Strain MHOM/CO/88/UA316 is L. guyanensis based on MLEE, but L. panamensis based on MLSA (Table 1).
For each strain, the number of correct typings at species, species complex, and subgenus level are reported. In addition, the incorrect species designations are indicated, some of which identified the wrong species in the correct complex (purple bars), others placing an isolate in the wrong complex (red bars). The strain identification by WHO code (Table 1) is given with the abscissa. Species, complexes, and subgenera are represented on top, with an indication of the New or Old World strain origin.