Figure 2. (Key Figure). Divergent Transcription at Enhancer Sequence.
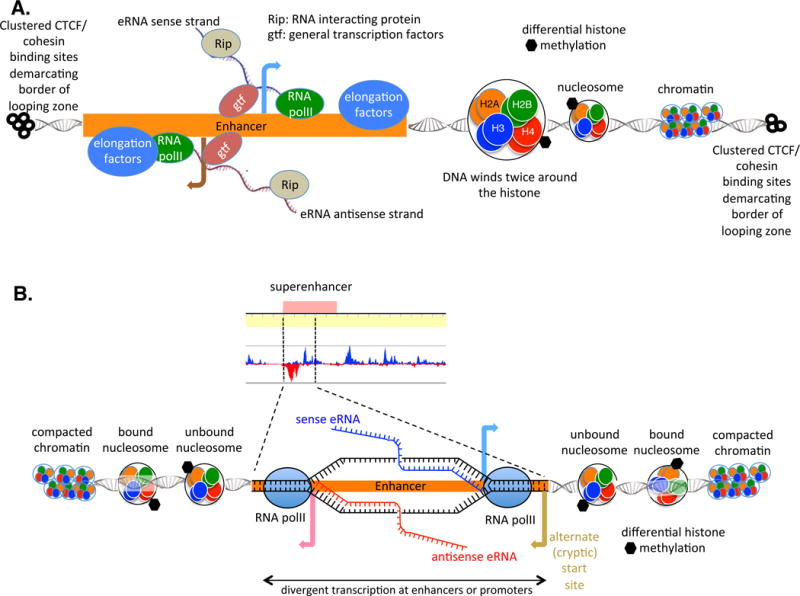
(A) Sense and antisense eRNA transcripts generated from an enhancer element. Transcripts may loop around and affect gene regulation by binding to promoters, as modeled in Figure 1. Note the presence of the clustered CTCF- and cohesin-binding sites, which demarcate the TAD. Looping between enhancers and promoters is believed to occur within a TAD but not between TADs. Looping occurs in open chromatin so that as distance from the enhancer increases, histones are compacted into nucleosomes and then further compacted into heterochromatin. Superenhancers are defined by altered histone methylation/acetylation. (B) In a superenhancer (series of enhancers in close proximity to one another causing high enhancer density), transcription of sense and antisense eRNAs in opposite orientations (by divergent enhancers) may mark the site for the cellular machinery as a location to which looping occurs. The antisense transcript may stall due to various hindrances, causing ‘flap-back’ of the nascent RNA and generating RNA–DNA structures called R loops. R loops attract AID whose activity causes alterations in the DNA and may allow looping to other areas (AID not shown in figure). Alternate (or cryptic) start sites may likewise generate transcripts (transcript not portrayed in figure). Abbreviations: AID, activation-induced deaminase; CTCF, [CK14]; eRNA, enhancer RNA; gtf, general transcription factor; Rip, RNA-interacting protein; RNA polII; RNA polymerase II; TAD, topologically associated domain.
