Figure 3. High Resolution of Genome Structure and Looping.
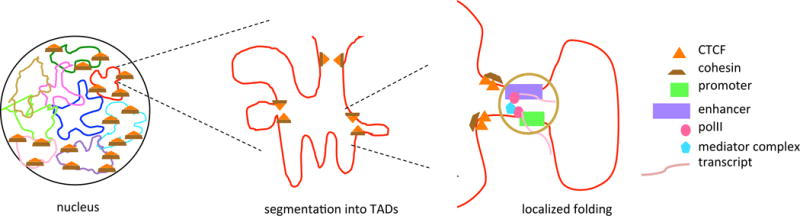
(A) Schematic of a mammalian nucleus with nine chromosomes indicated. Five of the chromosomes have TADs demarcated by CTCF/cohesin protein aggregates. (B) Magnified view of a TAD demarcated by CTCF/cohesin boundary marks. Note that subloops occur within the TAD – potentially bringing together a number of loops so that different promoters and enhancers interact with distant DNA sites concurrently (not indicated in figure) – but looping between TADs is thought not to occur except in states of disease. (C) Magnified view of the extrusion complex in B. Note the mediator complex interacting with the polymerases. Abbreviations: CTCF, [CK15]; TAD, topologically associated domain.
