Figure 4. Clustering of Sense–Antisense-Expressing Genes around Superenhancers.
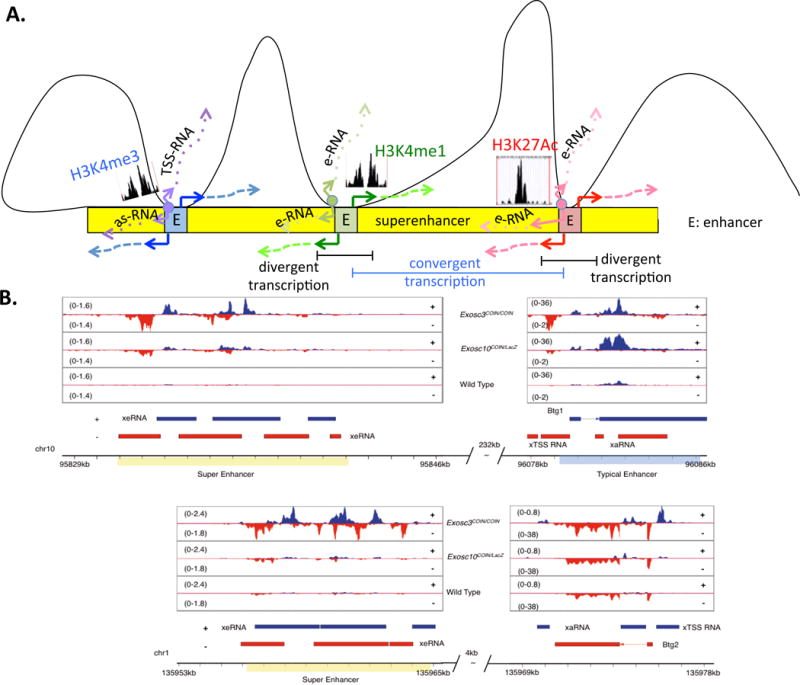
(A) A schematic showing (from left) a promoter (enriched for H3K4me3 marks and expressing TSS- and as-RNAs), (middle) an enhancer (enriched for H3K4me1 marks and expressing bidirectional enhancer RNAs), and (right) a superenhancer (enriched for H3K27Ac marks and expressing bidirectional enhancer RNAs) interacting with three separate enhancer loci within a superenhancer locus (blue, green, and red loci within the yellow superenhancer). Bidirectional transcription marks each of the three loci, potentially marking them as candidates for interaction with the superenhancer locus. Divergent transcripts are transcribed in opposite orientations (highlighted in black bar) but two closely-spaced divergent transcripts may form a situation of convergent transcription (highlighted in blue bar). (B) IGV[CK16] reads from cultured splenocytes illustrate that synthesis of as-RNAs may permit functional engagement with superenhancer elements to form higher-order chromosomal structures. (Top) A superenhancer–enhancer (overlapping the Btg1 gene) pair separated by a distance of 232 kb, expressing exosome-linked se-RNAs and TSS-RNAs, respectively. (Bottom) A superenhancer–enhancer (overlapping the Btg2 gene) pair separated by a distance of 4 kb, expressing exosome-linked se-RNAs and as-RNAs, respectively. Unshown: in examples where the enhancer–superenhancer are separated by distances >310 kb, the enhancer is less likely to express exosome-linked TSS-RNAs. Adapted from [12]. Abbreviations: as-RNA, antisense RNA; E, enhancer; eRNA, enhancer RNA; se-RNA, superenhancer RNA; TSS-RNA, transcription start site RNA.
