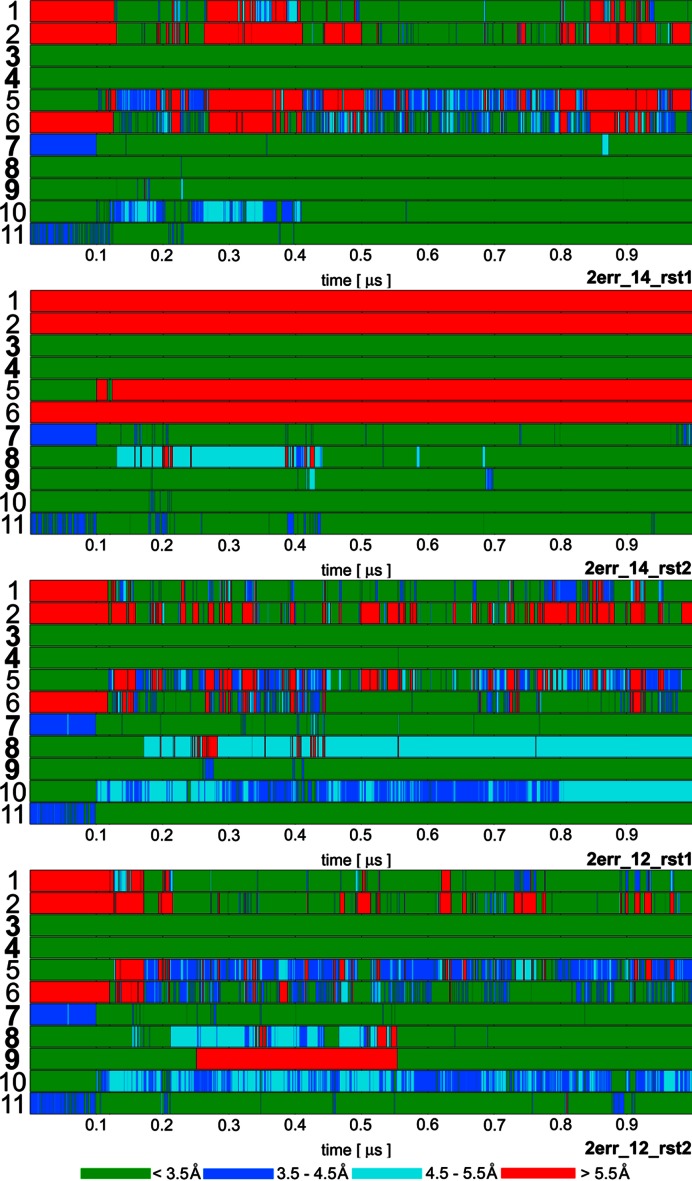
Figure 2.
Time development of heavy atom distances of specific intermolecular H-bond interactions in selected Fox-1 protein / RNA complex (PDB: 2err) simulations: 1. U1(N3)/Ser155(O); 2. U1(O2)/Asn151(ND2); 3. G2(N1)/Ile124(O); 4. G2(N2)/Ile124(O); 5. C3(N3)/Asn151(ND2); 6. C3(N4)/Ser155(O); 7. U5(N3)/Asn190(O); 8. U5(O2)/Thr192(N); 9. G6(N1)/Thr192(O); 10.G6(O6)/Arg118(sc); 11. G6(N7)/Arg118(sc). The H-bonds written in bold are present in the experimental NMR ensemble structures with over 90% occurrence. The H-bonds ‘1’ and ‘2’ are absent in the NMR ensemble but are often formed during the simulations. The ‘sc’ abbreviation for Arginine indicates any of the three side-chain nitrogen atoms potentially acting as donors in an H-bond. The bond angles were monitored to verify that short interatomic distances correspond to H-bonding but are not shown for space reasons. The remaining simulations are summarized in Supplementary Figure S4.