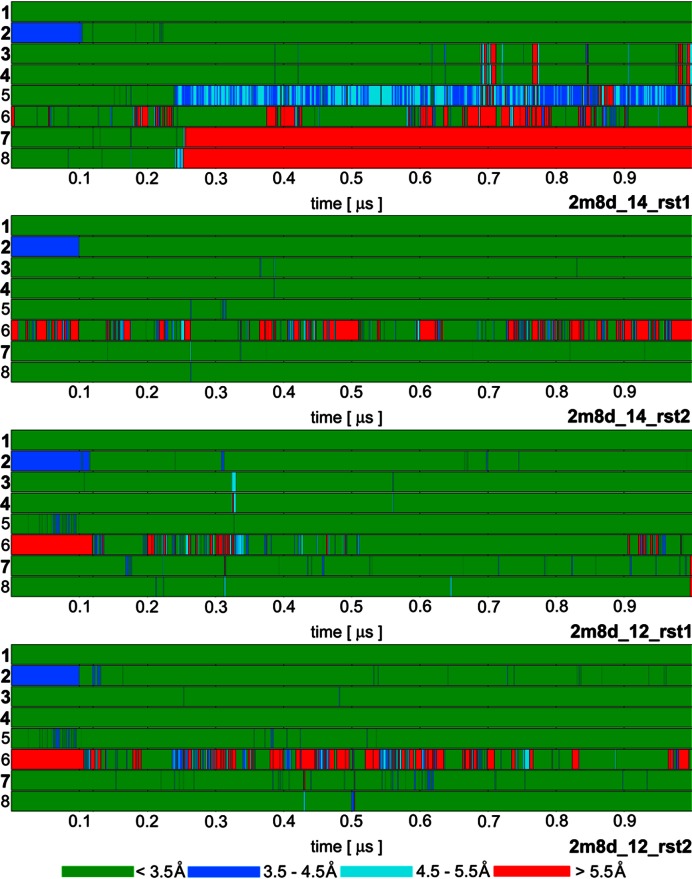
Figure 3.
Time development of heavy atom distances of specific intermolecular H-bond interactions in selected SRSF1 protein / RNA complex (PDB: 2m8d) simulations: 1. G5(N1)/Ala150(O); 2. G5(O6)/Ala150(N); 3. G6(N1)/Asp139(OD); 4. G6(N2)/Asp139(OD); 5. G6(O4')/Gln135(NE2); 6. G6(O6)/Arg142(sc); 7. A7(N6)/Asp136(OD); 8. A7(N1)/Ser133(OG). The H-bonds written in bold are present in the experimental NMR ensemble structures with over 90% occurrence. The H-bond ‘6’ is present in just one structure of the NMR ensemble but it is frequently observed in our simulations. The remaining simulations are summarized in Supplementary Figure S3.