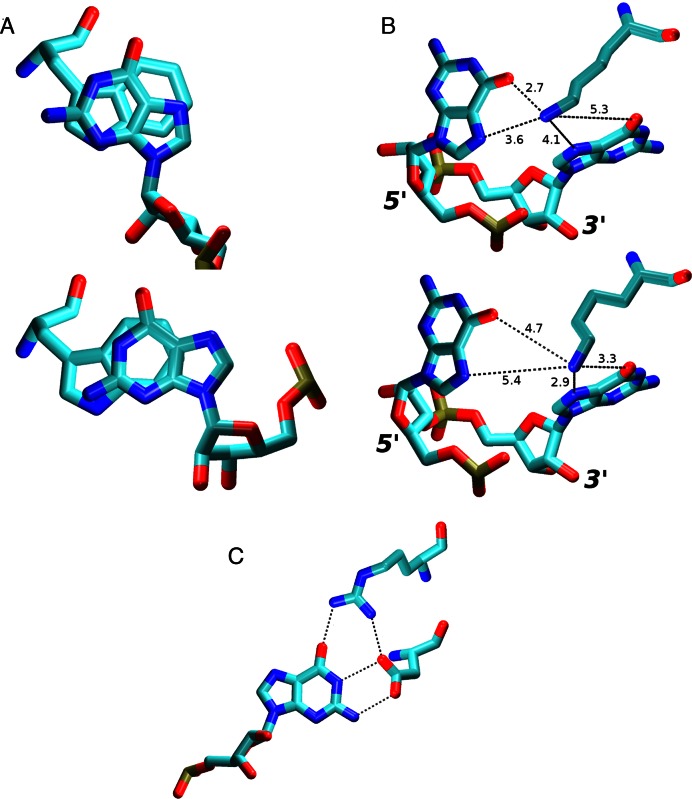
Figure 6.
(A) Overlap of G5 and Trp134 aromatic rings in the NMR (top) and in the simulations (bottom). This change, while minor, usually resulted into at least one G5/Trp134 NOE distance violation greater than 1 Å. (B) In simulations of the SRSF1 complex, the Lys138 side chain fluctuated between G5 (top) and G6 (bottom) Hoogsteen base edges. The typical heavy atom distances are shown (in Å). (C) The Arg142 side chain was often simultaneously interacting with G6 and Asp139 residues in the SRSF1 simulations, effectively increasing the protein's specificity for the guanine in this position by simultaneously recognizing the entire Watson-Crick edge of the base in a highly specific way.