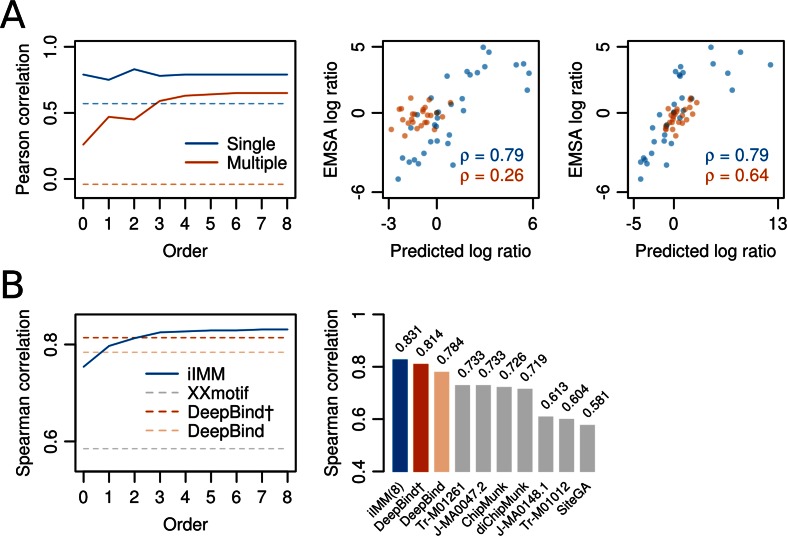
Figure 4.
Higher-order BaMMs boost accuracy of binding affinity predictions for weak sites. (A) Left: Pearson correlation between 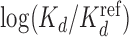 values for Klf4 binding to singly and multiply mutated consensus sites and
values for Klf4 binding to singly and multiply mutated consensus sites and 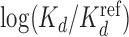 values predicted with models trained on Klf4-bound sequences from ChIP-seq. Solid lines: BaMM models; dashed lines: PWMs from XXmotif. Middle:
values predicted with models trained on Klf4-bound sequences from ChIP-seq. Solid lines: BaMM models; dashed lines: PWMs from XXmotif. Middle: 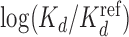 values measured by competitive EMSA assay versus values predicted by zeroth-order BaMM trained on ChIP-seq data. Right: same but predictions from fifth-order BaMM. Affinities to weak, multiply mutated sites (orange) are very badly predicted using a PWM (correlation 0.26) but decently using a fifth-order BaMM (correlation 0.64). (B) Left: same as (A), but showing Spearman rank correlations for FoxA2 binding affinities measured for 64 putative binding sites. DeepBind† and DeepBind differ only in model length (16 versus 24 bp). Right: Spearman correlations of our eighth-order BaMM and various other methods, adopted from Alipanahi et al. (50).
values measured by competitive EMSA assay versus values predicted by zeroth-order BaMM trained on ChIP-seq data. Right: same but predictions from fifth-order BaMM. Affinities to weak, multiply mutated sites (orange) are very badly predicted using a PWM (correlation 0.26) but decently using a fifth-order BaMM (correlation 0.64). (B) Left: same as (A), but showing Spearman rank correlations for FoxA2 binding affinities measured for 64 putative binding sites. DeepBind† and DeepBind differ only in model length (16 versus 24 bp). Right: Spearman correlations of our eighth-order BaMM and various other methods, adopted from Alipanahi et al. (50).