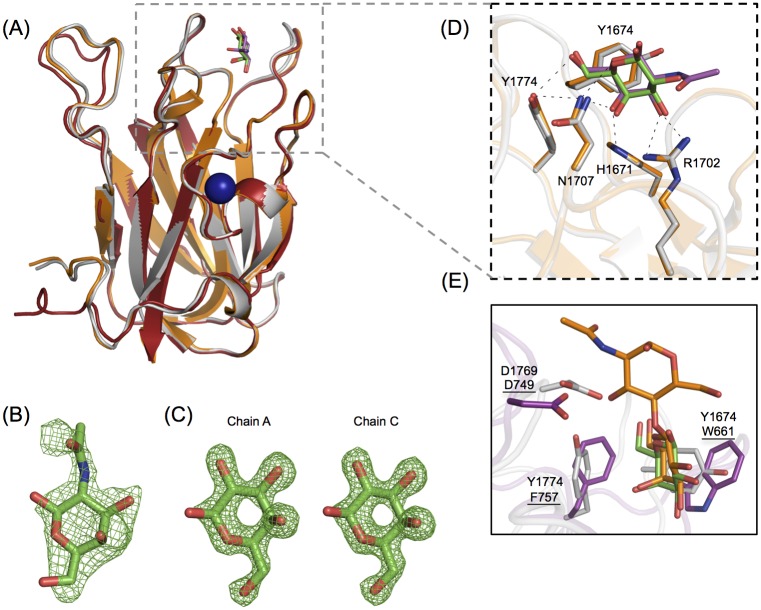
Fig 4. CpGH31 CBM32-3 recognizes GalNAc with the same set of residues employed by canonical galactose-binding CBM32s.
(A) Backbone cartoon structure overlay of the X-ray crystal structures of CBM32-3 in the apo-form (red) and in complex with galactose (grey) and GalNAc (orange), determined to 1.58 Å, 1.48 Å and 2.50 Å, respectively. Galactose is shown in green, and GalNAc in magenta. The Ca2+ ion observed in all three structures is depicted as a blue sphere. (B) Electron density of GalNAc bound to the single peptide chain of the CBM32-3:GalNAc structure, with the Fobs-Fcalc electron density map contoured to 2.5 σ. (C) Galactose bound to monomer chains A and C in the CBM32-3:galactose complex structure, with the Fobs-Fcalc electron density map contoured to 3.0 σ. (D) Expanded view of structural overlay of the ligand-coordinating residues from the galactose-bound (grey) and GalNAc-bound (orange) structures from (A). The same set of residues is involved in binding both ligands via hydrogen bonds (shown as black dashes). (E) A structural overlay of CpGH31 CBM32-3 (grey) bound to galactose (green, coordinating residues denoted) with CpGH84C CBM32 (magenta, coordinating residues underlined) bound to LacNAc (orange, PDB code 2J1E; [22]).