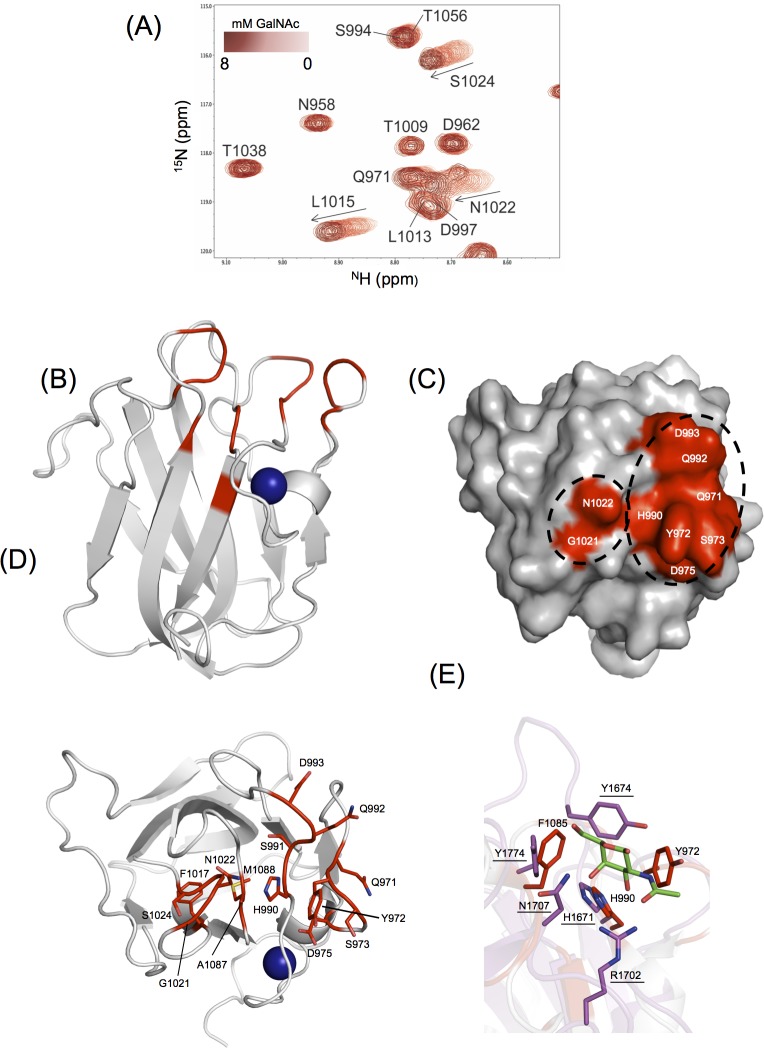
Fig 5. Identification of the CpGH31 CBM32-1 GalNAc binding site.
(A) Region of overlaid 1H-15N HSQC spectra of 500 μM CBM32-1 with increased amounts of GalNAc. (B) Backbone cartoon representation of the X-ray crystal structure of apo-CBM32-1 with residues whose backbone amide resonances were significantly perturbed (>1 standard deviation above the mean chemical shift change) in the presence of GalNAc shown in red. The calcium ion is shown as a blue sphere. (C) Surface representation of apo-CBM32-1 on which those significantly perturbed backbone amide resonances are displayed in red and identified in white as single-letter amino acid code. (D) Amino acid residues of CBM32-1 (shown as sticks and identified by single-letter code and position in CpGH31 sequence) whose backbone amide resonances display significant GalNAc-induced chemical shift changes are coloured red on a backbone cartoon representation of apo-CBM32-1. (E) Comparison of the GalNAc binding sites of CpGH31 CBM32-1 (grey, select residues denoted) and CpGH31 CBM32-3 (magenta, residues underlined) reveal structural conservation of Phe1985 and His990 with Tyr1774 and His1671, respectively. Tyr972 and Tyr1674 are located in different variable loop regions but are similarly positioned in the binding site.