Abstract
MicroRNAs (miRNAs) are a group of endogenous non-coding small RNAs that have critical regulatory functions in almost all known biological processes at the post-transcriptional level in a variety of organisms. The oriental fruit moth Grapholita molesta is one of the most serious pests in orchards worldwide and threatens the production of Rosacea fruits. In this study, a de novo small RNA library constructed from mixed stages of G. molesta was sequenced through Illumina sequencing platform and a total of 536 mature miRNAs consisting of 291 conserved and 245 novel miRNAs were identified. Most of the conserved and novel miRNAs were detected with moderate abundance. The miRNAs in the same cluster normally showed correlated expressional profiles. A comparative analysis of the 79 conserved miRNA families within 31 arthropod species indicated that these miRNA families were more conserved among insects and within orders of closer phylogenetic relationships. The KEGG pathway analysis and network prediction of target genes indicated that the complex composed of miRNAs, clock genes and developmental regulation genes may play vital roles to regulate the developmental circadian rhythm of G. molesta. Furthermore, based on the sRNA library of G. molesta, suitable reference genes were selected and validated for study of miRNA transcriptional profile in G. molesta under two biotic and six abiotic experimental conditions. This study systematically documented the miRNA profile in G. molesta, which could lay a foundation for further understanding of the regulatory roles of miRNAs in the development and metabolism in this pest and might also suggest clues to the development of genetic-based techniques for agricultural pest control.
Introduction
MicroRNAs (miRNAs) are non-coding small RNAs and are usually 21–24 nucleotide (nt) in lengths [1,2]. Accumulative reports suggest that miRNAs function as important gene expression regulators in almost all known physiological and pathophysiological process in varieties of eukaryotes and viruses [3–5]. Through the specific recognition between the miRNA seed region (nucleotides 2–8 from the 5’ en≈d) and the target sequences of mRNA existing in the 3’, 5’–untranslated region (3’UTR, 5’UTR) or the open reading frame, the finely tuning results of miRNAs normally lead to the degradation [6], transcriptional or translational repression [7,8] or even transcriptional enhancement of the target mRNA [9,10].
Since miRNA lin-4 was firstly discovered in Caenorhabditis elegans for regulation of development timing [11], with the development of next generation sequencing platforms, thousands of miRNAs have been identified in human, animals, plants, and viruses [12] and are available on miRBase (http://www.mirbase.org/). Until now, 35828 mature miRNAs have been identified from 220 species, in which 26 species belong to insects (http://www.mirbase.org/). Due to the absence of complete genome sequences, whereas, most of the available sequences of insect miRNAs were determined from model organisms, including 15 species of Diptera (12 Drosophila, Aedes aegypti, Anopheles gambiae and Culex quingquefasciatus), 4 species of Hymenoptera (Apis mellifera and 3 Nasonia), 4 species of Lepidoptera (Bombyx mori, Manduca sexta, Heliconius melpomene and Plutella xylostella), 1 Hemiptera species (Acyrthosiphon pisum), 1Orthoptera species (Locusta migratoria), and 1 Coleoptera species (Tribolium castaneum).
miRNA play various regulatory roles in insects, mainly in developmental regulations of germ cell line [13], wing formation [14,15], muscle development [7], neurogenesis [16], apoptosis [17], phenotypic plasticity [18,19], reproduction [18], metamorphosis [20], behavior [21,22], stress resistance [23] and host-pathogen interaction [24,25]. For the multiply regulatory feature, miRNAs are usually expressed in a precisely temporal and spatial pattern [26–29]. Thus, accurate quantifications of miRNA profiles in specific experimental conditions are essential to their functional detection. Up to date, three technologies, microarray [30], bead-based flow cytometry [31], and quantitative real time PCR (qPCR) [32,33] have been developed to study expressional profiles of miRNAs. qPCR, for its sensitivity, flexibility and convenience, has been widely applied to measure miRNA expression levels in diverse organisms. A large number of studies have revealed that appropriate normalizers are critical to the accuracy of gene expression evaluation [34–36].
The oriental fruit moth, Grapholita molesta (Busck) (Lepidoptera: Tortricidae) is a worldwide pest of stone and pome fruits in most temperate fruit-growing regions [37] and prefers the new shoots and fruits of host plants within the family Rosacea for boring and internal-feeding [38,39]. Grapholita molesta normally has three to five generations per season. The adults migrate from peach orchards to pear or apple orchards. Therefore, outbreaks often occur in the mixed-planting orchards, which result in substantial losses in yields and incomes [40,41]. Nowadays, the main management method is to use insecticides for egg and neonate control. Unfortunately, insecticide resistance has occurred in G. molesta [42]. An auxiliary technique, pheromone-based mating disruption of adults, is effective but with high management costs [43]. A better understanding about the regulatory mechanisms of the metabolism, development and metamorphosis of G. molesta would shed light on the development of new techniques for efficient management of this borer pest.
In the present study, on the basis of the Illumina small RNA (sRNA) library sequencing platform, the miRNA transcriptome of G. molesta was sequenced and performed with systematically in silico analysis in reference to the transcriptome of G. molesta and the genome of Danaus plexippus. We also validated the expressional stability of the candidate reference genes and made selection of suitable normalizers under different biotic and abiotic experimental conditions. Our results would provide useful information for revealing the miRNA-involved regulatory mechanism of the development and metabolism in G. molesta and would also supply a reference for deeper understanding of the expressional profile and regulatory function of miRNA in other non-model insects and agricultural insect pests.
Materials and methods
Ethics statement
In this study, the larvae of oriental fruit moth G. molesta were originally collected in the Institute of Pomology in Liaoning province, China. No permissions were required for the insect collection, as the orchards are experimental plots belonging to the Institute of Pomology in Liaoning province. The “List of Protected Animals in China” excludes insects.
Insect rearing
Grapholita molesta used in this study were reared under laboratory conditions (26 ± 1°C, 70 ± 10%, 15 h L: 9 h D) on fresh apples and artificial diet over four years as our previous study [44]. Different instars of G. molesta were carefully picked out from rotten apples with soft tweezers. Five larvae of the same instar were reared together in one finger-shaped glass tube (5.5 cm in length ×2.2 cm in diameter) on artificial diet jelly. After pupation, pupae were collected from the cotton-wool tampon on the top of the tubes, and newly-emerged adults were transferred to beakers (1 L in volume) with fresh apples inside for egg laying. Cotton balls soaked with 10% honey solution were provided for nutrient supplement.
Sample collection and total RNA extraction for sRNA library construction
Healthy samples of G. molesta, including eggs, the first to fifth instars, prepupae, pupae, female and male adults were collected and snap-frozen in liquid nitrogen. Three hundred eggs, one hundred first and second instars and ten pooled frozen insects of other developmental stages were respectively ground into powder in liquid nitrogen with cooled mortars and pestles. Aliquots of the powdered samples were separately transferred into 1.5 ml RNase-Free Eppendorf tubes. The total RNA samples were isolated with Trizol reagent (Takara, Kyoto, Japan) according to the instruction. The quality and quantity of RNA extracts were then measured with Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA) and Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA).
sRNA library construction and sequencing
RNA extracts from each stage of G. molesta were mixed in an equal amount and then 1 μg of the resulting RNA mix was used for construction of sRNA library using TruSeq Small RNA Sample Prep Kit (Illumina, San Diego, CA, USA). Briefly, sRNAs of 15–30 bp were fractionated on 15% PAGE (polyacrylamide gel electrophoresis); 5’ and 3’ RNA adaptors were then ligated to the recovered sRNA; cDNAs were finally obtained after reverse transcription. After PCR amplification, 6% polyacrylamide TBE (Tris-borate-EDTA) gel purification and cluster formation, the resulting cDNAs were subjected to the single-end sequencing for 50 cycles (1 bp reads per cycle) on an Illumina Hiseq2500 (Illumina, San Diego, CA, USA) in LC-BIO (Hangzhou, China).
Sequence filter and annotation
The raw reads were subjected to the Illumina pipeline filter (Solexa 0.3) for quality assessment. Clean reads were then obtained through data process with the program ACGT101-miR (LC Sciences, Houston, Texas, USA) after removing adapter dimers, junk reads and common non-coding RNA families (rRNA, tRNA, snRNA, snoRNA). Finally, the unique reads were screened out by eliminating the repeated sRNAs from the clean reads database with single sequence left.
To identify miRNAs, the unique sequence dataset with length of 18–26 bp were mapped in the miRBase (release 21, http://www.mirbase.org/) by BLAST search, and length variations at both 3’ and 5’ ends and one mismatch inside the sequence were allowed in the alignment. The unique reads that mapped to the known miRNAs and/or pre-miRNAs of specific species were determined to be conserved miRNAs. The remaining sequences were further mapped against the transcriptome of G. molesta (unpublished data) and genome of highly homological lepidopteran D. plexippus (downloaded from MonarchBase website: http://monarchbase.umassmed.edu). The reads that could map to the transcriptome and/or genome were further subjected to the secondary hairpin prediction using RNAfold software (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). The sequences of transcriptome/genome that 20 bp upstream and 60 bp downstream of the mapped sequences and the sequences 60 bp upstream and 20 bp downstream of the mapped sequences were both extracted as candidate precursors for secondary structure prediction. The criteria for secondary structure prediction were: (1) number of nucleotides in one bulge in stem (≤ 12); (2) number of base pairs in the stem region of the predicted hairpin (≥ 16); (3) cut of free energy (kCal/mol ≤ 15); (4) length of hairpin (up and down stems + terminal loop ≥ 50); (5) length of hairpin loop (≤ 20); (6) number of nucleotides in one bulge in mature region (≤ 8); (7) number of biased errors in one bulge in mature region (≤ 4); (8) number of biased bulges in mature region (≤ 2); (9) number of errors in mature region (≤ 7); (10) number of base pairs in the mature region of the predicted hairpin (≥ 12); (11) percent of mature in stem (≥ 80%). The reads with precursors that satisfied all of the above standards and with more than 10 reads account were determined as novel miRNAs. The secondary structure of novel miRNAs were then constructed using srna-workbench V2.5.0 (http://srna-workbench.cmp.uea.ac.uk/downloadspage/) with the precursor sequence and the brief-code of the corresponding secondary structure of each novel miRNA.
Validation of the Illumina sequencing
To validate the Illumina sequencing, 10 conserved miRNAs that have been reported with relative stable expression in other organisms [45–47] and 10 novel identified miRNAs with high abundance in sRNA library of G. molesta (S1 Table) were selected and their expression level in the sample for construction of sRNA library were checked using qPCR. What’s more, the 20 selected miRNAs were respectively cloned into pMD18-T (Takara) and were further validated with Sanger sequencing by Beijing Genomics Institute (BGI).
Cluster and conservation analysis
Cluster analysis was conducted among miRNAs at the distance of 3 kb, 5 kb, 10 kb and 50 kb within the genome of D. plexippus. One cluster was determined when at least two miRNAs could be found in a specific distance range.
The miRNA sequences of arthropods, C. elegans and Homo sapiens have been downloaded from miRBase (release 21). Conservation of miRNA families among G. molesta and other arthropods was compared through BLASTn analysis. Phylogenetic relationships among 27 insect species belonging to 6 orders in Hexapoda and 4 species in the other three phyla of arthropods have been analyzed. The topology tree was constructed according to previous reports [48,49].
Target prediction and KEGG enrichment analysis
Putative target genes of miRNAs in G. molesta were concurrently predicted by TargetScan 50 and miRanda 3.3a through identification of miRNA binding sties on the 3’- UTR sequences of G. molesta transcriptome. The target genes with context score percentile less than 50 were then eliminated by TargetScan. The ones with max energy greater than -10 were screened out by miRanda. Finally, the target genes simultaneously recommended by the two algorithms were defined as the final prediction.
Functional pathway determination of target genes was performed through the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis against the public database (http://www.genome.jp/kegg) [50]. Pathways with P–value of fisher’s exact test ≤ 0.05 were determined to be significantly enriched pathways for metabolism or signal transduction. For elucidation of the interactions between miRNAs and target genes in metabolisms and signal transductions, networks of miRNA-KEGG in G. molesta were further analyzed with miRNA-mRNA target sequence analysis technique and were descripted with Cytoscape 2.8.3.
Sample preparation and candidate sRNA selection for reference gene selection
Eight experimental conditions had been considered for study on expressional stability of sRNAs in G. molesta, including two intrinsic biotic conditions (developmental stage and tissue) and six exogenous stress conditions (temperature, photoperiod, starvation, JH injection, dsRNA injection and insecticide).
For samples of different developmental stages, 300 eggs, 200 first instars, 30 second instars, 10 third instars, 10 fourth instars, 7 fifth instars, 10 prepupae, 10 female pupae, 10 male pupae, 10 female adults (3 d old post-emergence) and 10 male adults (3 d old post-emergence) of G. molesta have been collected respectively as one replicate. For sample preparation of different tissues, 20 fifth instars (2 d old post-molt) and 10 adults (3 d old post-emergence) were dissected under a binocular microscope in 10 mM cold phosphate buffered saline (PBS, pH 7.8) as one replicate; after rinsed in PBS, samples of each tissue, including 9 tissues of larvae (head, cuticle, foregut, midgut, hindgut, Malpighian tubule, fatbody and ventral nerve cord) and 4 tissues of adults (head, thorax, abdomen and leg) were obtained respectively from pooled dissections.
For sample preparation of G. molesta subjected to stresses of varying temperature, photoperiod and food deprivation, the third instars were treated under nine temperature conditions (4, 26 and 40°C for 2, 12 and 24 h, respectively), eight photoperiod conditions (24 h L: 0 h D, 0 h L: 24 h D, 14 h L: 10 h D and 10 h L: 14 h D for 1 d and 2 d, respectively), and four starvation conditions (starved for 12 h, 24 h, 48 h and 48 h followed by 24 h refeeding). For insect samples pretreated with double strand RNA (dsRNA) and juvenile hormone (JH) injection, the fifth instars were respectively injected with artificially synthesized dsRNA of met (encoding putative JH receptor Methoprene-tolerant) and chemical reagent JH III (Sigma-Aldrich, Milwaukee, WI, USA). DsRNA of egfp (encoding Enhanced Green Fluorescent Protein) and chemical reagent dimethyl sulphoxide (DMSO, solvent of JH III, Sigma-Aldrich, Milwaukee, WI, USA) were injected as control treatments accordingly. DsRNAs of met and egfp were artificially prepared with MEGAscript RNAi kit (Ambion) with primer pairs listed in S2 Table. For stress induced by insecticide, the third instars were topically applied with LD50 of 2.5% β-cypermethrin (1 μL of 1500 times dilution per larva, or 0.625 ng per larvae) calculated using PoloPlusTM software (LeOra software, Berkeley, CA, USA) and were then sampled, respectively, at 12 h, 24 h and 48 h post application. Ten insects pretreated under each exogenous stress condition were collected as one replicate. Three replicates were collected for all of the eight treatments.
All insect samples were snap frozen in liquid nitrogen and stored at -80°C. Total RNA of each sample was then extracted with Trizol reagent as above and checked with Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, cDNAs of sRNAs were synthesized after reverse transcription from the qualified RNA with A260/280 value of 1.8–2.3 and A260/230 value ≥ 2.0 using miScript II RT Kit (Qiangen, Dusseldorf, Germany). Universal reverse primer (GAATCGAGCACCAGTTACGC) was used for cDNA synthesis according to the manufacture’s protocol. All of the resulting cDNA samples were stored at −20°C for sRNA expressional analysis.
For reference RNAs selection, 12 sRNAs, including two commonly used normalizer (small nuclear RNA U6 and small ribosomal RNA 5SrRNA) for miRNA quantification in mammals and plants [51–54] and 10 miRNAs (bmo-miR-2b-3p_1ss22GC, pxy-mir-6497-p3_1ss7CT, hsa-miR-16-5p, bmo-miR-281-3p_L-2R+2, bmo-miR-279a_R+2, bmo-miR-9a-5p, bmo-miR-998_R+2, bmo-miR-305-5p_R+1, bmo-let-7-5p and mse-miR-92a) which have been used for normalization in previous reports [45–47] and showed abundant expression in the sRNA library of G. molesta have been selected as candidate reference genes for miRNA profile study of G. molesta.
Quantitative PCR analysis
qPCR with high sensitivity and specificity was adopted for validation of sRNA library sequencing and quantification of selected candidate reference genes through measurement of mature sRNA expression.
The forward primers for qPCR amplification of miRNAs were designed according to the specific sequences of miRNAs with some modifications at 5’ ends for adjustment of GC contents (S1 Table). The reverse primer for qRT-PCR analysis was the universal primer used for cDNA synthesis above. All primers were synthesized commercially (Sangon Biotechnology, Shanghai, China) and diluted to 10 μM. qPCR was conducted on a Bio-Rad CFX ConnectTM Real-Time PCR Detection System (Hercules, CA, US) using miScript SYBR® Green PCR kit (Qiagen). The amplification was performed following the program as: 94°C for 15 min for denaturation, 40 cycles of 95°C for 15 s, 55°C for 30 s, 72°C for 30 s for collection of amplification signal and 70°C for 30 s for reaction termination. The dissociation protocol was also carried out for melting curve analysis on the specific amplification. The amplification efficiency (E%) and correlation coefficient (R2) for amplification of each sRNA were calculated according to the standard curve generated from the five 5-fold serial dilution points of cDNAs and their corresponding Cq values [35,55].
Statistical analysis for selection and validation of reference genes
Global mean that was defined as the mean Cq value of all expressed sRNAs in a given sample [34] was calculated. The Spearman’s rank correlation coefficient rs between Cq value of each candidate reference gene and the corresponding global mean was then respectively analyzed under each experimental condition [56]. The expressional stabilities of the candidate reference genes were preliminarily ranked according to their rs values and the sRNA with rs value closer to 1 was considered more stable.
The expressional stability of candidate sRNAs with rs value > 0 were further assessed using commonly used software geNorm [57]. The linear scale expression quantity of each candidate gene was calculated by the equation 2(-ΔCq) (according to the handbook of geNormTM kit with perfect probe, PrimerDesign Ltd.) and loaded in the Excel-based geNorm software. The optimal number of reference genes used under each experimental condition was also recommended by geNorm through pairwise variation analysis.
To validate the selected reference genes, transcriptional level of let-7 in G. molesta at different developmental stages were detected and compared with different normalizers, including the single one best recommendation hsa16 (NF1), the recommended combination hsa16 + U6 + bmo2b (NF(1–3)), one least stable miRNA bmo281 (NF8) ranked by geNorm, and single mse92a (NF12) with the lowest rs value. Relative expression levels of let-7 were calculated according to the ΔΔCq method. The geometric mean calculated from the Cq values of the three sRNAs (hsa16, U6, bmo2b) were used as normalization factor for NF(1–3). For each specific developmental stage, the significant difference among the expression levels of let-7 calculated according to different normalization factors were statistically analyzed using one-way ANOVA with SPSS Statistics 17.0 (SPSS Inc., Chicago, IL, USA). Specifically, Tukey test was adopted for analysis of gene expression in G. molesta of fifth instar, male pupa, female pupa, and female adul. Games-Howell test was performed in analysis of gene expression in G. molesta of first instar, second instar, fourth instar and prepupa. For expressional analysis in G. molesta of third instar and male adult, Games-Howell test was also applied after square root transformation of the relative gene expression values.
Results
Profile of sRNA library
In order to understand the miRNA profile, a sRNA library was constructed with a mixture of RNA extracted from G. molesta at varying developmental stages. A total of 16,305,575 raw reads were obtained, including 2,219,608 unique sequences. After trimming of the adaptor and junk reads, filtering out the Rfam reads (rRNA, tRNA, snoRNA, snRNA and other Rfam RNA), and removing the repeats, a total number of 5,998,101 unique reads were finally obtained for further analysis (Table 1).
Table 1. Overview of sRNA sequencing data.
| Category | Total reads | Percentage of total reads (%) | Unique sequence | Percentage of unique sequence (%) |
|---|---|---|---|---|
| Raw reads | 16,305,575 | 100.00 | 2,219,608 | 100 |
| 3'adaptor & length filtera | 9,804,065 | 60.13 | 1,660,906 | 74.83 |
| Junk readsb | 24,021 | 0.15 | 5,511 | 0.25 |
| Rfamc | 477,365 | 2.93 | 19,662 | 0.89 |
| rRNA | 328,485 | 2.01 | 12,880 | 0.58 |
| tRNA | 62,488 | 0.38 | 3,557 | 0.16 |
| snoRNA | 804 | 0.00 | 227 | 0.01 |
| snRNA | 2,343 | 0.01 | 555 | 0.02 |
| other Rfam RNA | 83,245 | 0.51 | 2,443 | 0.11 |
| Repeats | 5,190 | 0.03 | 456 | 0.02 |
| Unique reads | 5,998,101 | 36.79 | 533,252 | 24.02 |
a Length filter: reads with length <18 and >26.
b Junk reads: > = 2N, > = 7A, > = 8C, > = 6G, > = 7T, > = 10Dimer, > = 6Trimer, or > = 5Tetramer.
c Rfam: collection of many common non-coding RNA families (including rRNA, tRNA, snoRNA, snRNA and other Rfam RNA) except microRNA; http://rfam.janelia.org.
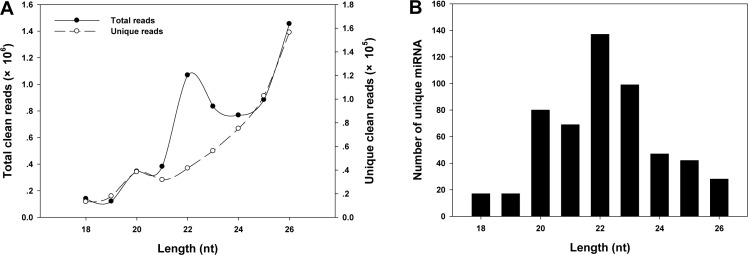
The length of sequenced sRNAs ranged from 18–26 nucleotides (nt). The length of the total clean reads mainly showed two peaks, with the highest one at 26 nt and the other at 22 nt (Fig 1A). The number of the unique clean reads increased with the lengths of the reads (Fig 1A), and the highest number of miRNAs were found with length of 22 nt (Fig 1B).
Fig 1. Length distribution of sRNAs in the sRNA library of G. molesta.
A, The total number of clean and unique reads of sRNAs in lengths of 18–26 nt; B, the number of unique miRNAs ranging from 18–26 nt.
Identification of conserved and novel miRNAs
After all of the unique clean reads were subjected to the alignment in the miRBase 21.0, transcriptome of G. molesta and the genome of D. plexippus, and were performed with the analysis of the precursors’ hairpin, a total of 536 mature miRNAs were obtained in the sequenced sRNA library of G. molesta (S3 Table). Among the defined miRNAs, 38.06% were mapped to the transcriptome of G. molesta (2.94% conserved and 97.6% novel) (S4 Table and S1 Fig), and 27.24% were mapped to the genome of D. plexippus (67.81% conserved and 32.19% novel) (S5 Table and S1 Fig). The identified 536 miRNAs were composed of 291 conserved and 245 novel miRNAs which were, respectively, belonged to 206 conserved and 230 novel seed-based miRNA families (S6 Table).
Among the 291 conserved miRNAs, only 40 miRNA showed high expression levels (> 2000 reads) (Table 2), and miR-10, miR-8, miR-276, miR-14 and miR-281 were identified as the most abundantly expressed conserved miRNAs in G. molesta. Majority of the conserved miRNA showed middle (132 miRNAs with 10–1999 reads) to low (119 miRNAs with < 10 reads) expressional levels (S4and S5 Tables). In order to detect the conservation of miRNAs among G. molesta and other arthropods, 79 miRNA families identified in the sRNA library of G. molesta were compared within 31 arthropods, and the phylogenetic relationship among these arthropods were constructed with C. elegans and H. sapiens as outgroup taxa (Fig 2). miRNAs were relatively conserved in the same order, and the miRNAs were more conserved in orders with closer phylogenetic relationships. Insects appeared having more expressed miRNAs than those in other arthropods, Nematoda and Chordata, and lepidopterans seemed owing the most abundant miRNAs in Hexapoda. In the present study, miR-9 was conserved in all of the insect species, and miR-745 was conserved in Lepidoptera but absent in other species.
Table 2. The most abundant conserved miRNAs in the sRNA library of G. molesta.
| miRNA | Sequence | Length | Seed | CG(%) | Free energy | Reads account |
|---|---|---|---|---|---|---|
| bmo-miR-10-5p_L+1 | TACCCTGTAGATCCGAATTTGT | 22 | ACCCTGT | 44.20 | -37.10 | 338,584 |
| mse-miR-8 | TAATACTGTCAGGTAAAGATGTC | 23 | AATACTG | 50.00 | -40.50 | 330,445 |
| bmo-miR-276-3p | TAGGAACTTCATACCGTGCTCT | 22 | AGGAACT | 44.90 | -38 | 125,388 |
| bmo-miR-14-3p_R+1 | TCAGTCTTTTTCTCTCTCCTAT | 22 | CAGTCTT | 40.00 | -31.70 | 55,077 |
| bmo-miR-281-5p_R+1 | AAGAGAGCTATCCGTCGACAGTA | 23 | AGAGAGC | 39.80 | -39.60 | 29,116 |
| bmo-miR-11-3p_1ss11AG | CATCACAGTCGGAGTTCTAGCT | 22 | ATCACAG | 56.50 | -38.80 | 27,252 |
| bmo-miR-279d-3p | TGACTAGATTTTCACTTATCCT | 22 | GACTAGA | 39.40 | -22.60 | 26,764 |
| bmo-miR-31-5p_L+1R+1_1ss9AT | AGGCAAGATGTCGGCATAGCTGA | 23 | GGCAAGA | 57.00 | -48.20 | 25,425 |
| bmo-mir-6497-p5 | GGAATAAGGATTGGCTCTGAGGAC | 24 | GAATAAG | 64.80 | -36.90 | 24,686 |
| mse-miR-263a_R-1 | AATGGCACTGGAAGAATTCACGG | 23 | ATGGCAC | 50.00 | -34.10 | 21,512 |
| bmo-miR-9a-5p | TCTTTGGTTATCTAGCTGTATGA | 23 | CTTTGGT | 40.40 | -33.30 | 21,174 |
| mse-miR-6094 | TATTCGAGACCTCTGCTGATCCT | 23 | ATTCGAG | 51.30 | -51.40 | 19,051 |
| bmo-miR-2b-3p_1ss22GC | TATCACAGCCAGCTTTGTTGACT | 23 | ATCACAG | 46.20 | -48.30 | 18,960 |
| bmo-miR-263a-5p_R+4 | AATGGCACTGGAAGAATTCACGGGA | 25 | ATGGCAC | 51.50 | -40.50 | 14,803 |
| bmo-miR-71-3p_L-1R+1 | TCTCACTACCTTGTCTTTCATG | 22 | CTCACTA | 47.70 | -44.50 | 13,455 |
| mse-miR-277 | TAAATGCACTATCTGGTACGACA | 23 | AAATGCA | 51.60 | -39.50 | 8,369 |
| bmo-miR-277-3p | TAAATGCACTATCTGGTACGACA | 23 | AAATGCA | 51.70 | -56.50 | 8,369 |
| bmo-miR-279b-3p_R-1 | TGACTAGATCTACACTCATTG | 21 | GACTAGA | 40.50 | -47.60 | 7814.50 |
| bmo-miR-282-5p_L-3R-2 | TAGCCTCTCCTTGGCTTTGTCT | 22 | AGCCTCT | 46.00 | -37.90 | 6,909 |
| bmo-miR-750-3p_R+2 | CCAGATCTATCTTTCCAGCTCA | 22 | CAGATCT | 48.80 | -33 | 6,646 |
| pxy-miR-274 | TTTGTGACCGTCACTAACGGGCA | 23 | TTGTGAC | 43.40 | -29.50 | 6,383 |
| bmo-miR-10-3p | CAAATTCGGTTCTAGAGAGGTTT | 23 | AAATTCG | 44.20 | -37.10 | 5,036 |
| bmo-miR-31-5p_L+1R-2_1ss9AT | AGGCAAGATGTCGGCATAGC | 20 | GGCAAGA | 57.00 | -48.20 | 4,591 |
| bmo-let-7-5p | TGAGGTAGTAGGTTGTATAGT | 21 | GAGGTAG | 52.00 | -50.90 | 4,349 |
| mse-miR-278 | TCGGTGGGATCTTCGTCCGTTT | 22 | CGGTGGG | 54.30 | -39 | 4,269 |
| bmo-miR-281-3p_L-2R+2 | TGTCATGGAGTTGCTCTCTTTA | 22 | GTCATGG | 39.80 | -39.60 | 4,260 |
| bmo-miR-252-5p | CTAAGTACTAGTGCCGCAGGAG | 22 | TAAGTAC | 37.20 | -33.10 | 4,031 |
| dme-miR-10-5p_L+1_1ss23TA | TACCCTGTAGATCCGAATTTGTA | 23 | ACCCTGT | 47.60 | -48.60 | 3,798 |
| bmo-miR-1175-3p_R-2 | TGAGATTCAACTCCTCCAACTT | 22 | GAGATTC | 38.20 | -23.50 | 3,749 |
| mse-mir-8-p5 | CATCTTACCGGGCAGCATTAGA | 22 | ATCTTAC | 50.00 | -40.50 | 3,279 |
| bmo-miR-7-5p_R+1 | TGGAAGACTAGTGATTTTGTTGTT | 24 | GGAAGAC | 34.70 | -24.30 | 3,037 |
| bmo-mir-6497-p3 | GCGTGTCGGGTTTGGACGGGAAG | 23 | CGTGTCG | 64.80 | -36.90 | 2,985 |
| dme-miR-8-3p_R+1 | TAATACTGTCAGGTAAAGATGTCC | 24 | AATACTG | 52.50 | -51.30 | 2,914 |
| bmo-miR-133 | TTGGTCCCCTTCAACCAGCTGT | 22 | TGGTCCC | 42.70 | -39.90 | 2,828 |
| bmo-miR-305-5p_R+1 | ATTGTACTTCATCAGGTGCTCTGG | 24 | TTGTACT | 56.80 | -48.30 | 2,595 |
| bmo-miR-263a-5p_R+5 | AATGGCACTGGAAGAATTCACGGGAA | 26 | ATGGCAC | 45.30 | -34 | 2,590 |
| bmo-miR-279c-3p_R+1 | TGACTAGATCCATACTCGTCTGC | 23 | GACTAGA | 40.20 | -36.90 | 2469.50 |
| bmo-miR-317-3p_L-2 | TGAACACAGCTGGTGGTATC | 20 | GAACACA | 57.00 | -41.50 | 2,318 |
| bmo-miR-2765 | TGGTAACTCCACCACCGTTGGC | 22 | GGTAACT | 57.60 | -59.80 | 2,154 |
bmo: Bombyx mori.
mse: Manduca sexta.
pxy: Plutella xylostella.
dme: Drosophila melanogaster.
Fig 2. Conservation analysis of 79 miRNA famililes within 33 species.
Conservation analysis of the 79 miRNA families identified from the sRNA library of G. molesta was conducted among 33 species belonging to the three phyla, Arthropoda, Nematoda and Chordata. The miRNA information of the other 32 species was retrieved from the miRBase. Colored box indicated the presence of the conserved miRNA family and the same color indicated species belonging to the same order of insect in Hexapoda, the same class in Arthropoda except for the Hexapoda species, C. elegans in Nematoda, and Homo sampiens in Chordata. The phylogenetic tree was constructed according to previous reports [48,49].
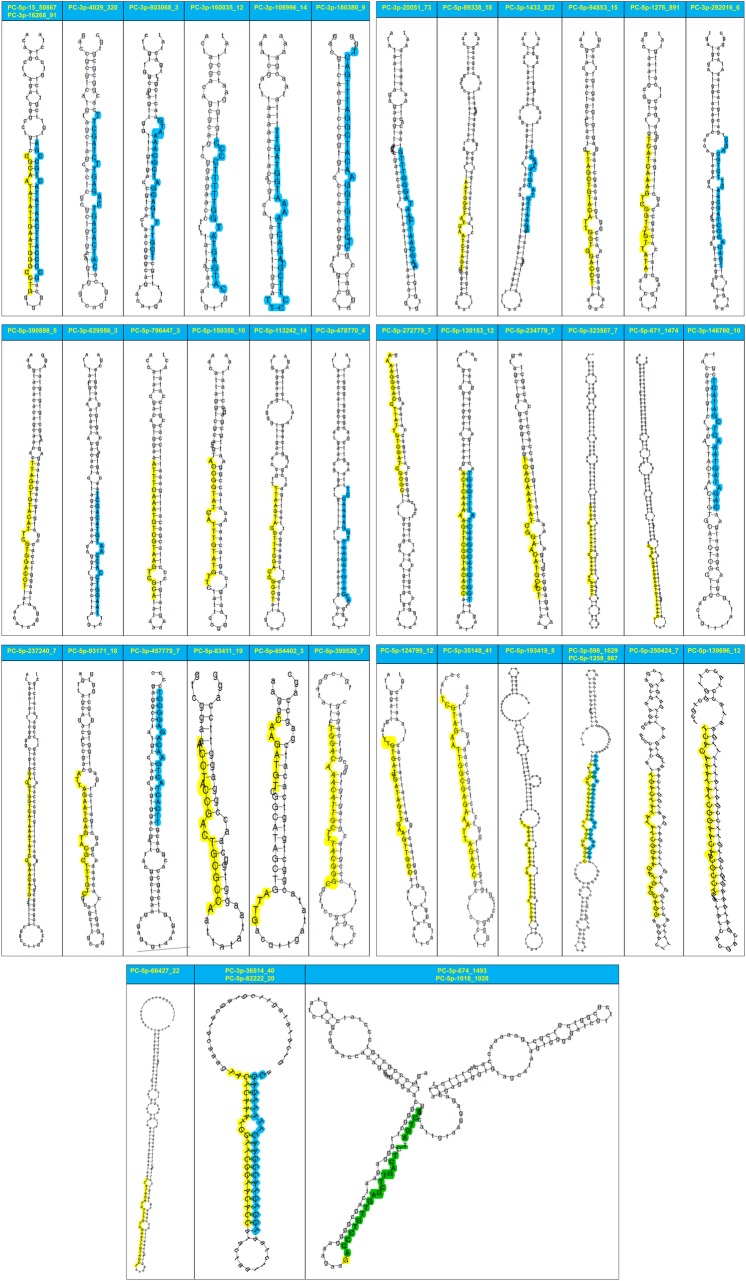
Most (82.04%) of the identified novel miRNAs showed low expressional levels (< 10 reads) (S4and S5 Tables), 39 (15.92%) of the novel miRNAs showed middle abundance (10–1999 reads) (Table 3), only 5 novel miRNAs exhibited with high abundance (> 2000 reads) (Table 3). The 5 highly expressed novel miRNAs were gmo-miR-PC-5p-15_50867 (127,819 reads), gmo-miR-PC-3p-598_1629 (17,872 reads), gmo-miR-PC-3p-674_1493 (17,424 reads), gmo-miR-PC-5p-1258_867 (14,307 reads) and gmo-miR-PC-5p-1018_1028 (9,152 reads), all of which accounted about 96.94% of all the identified novel miRNA reads. The secondary hairpin structures of the 44 novel miRNAs with middle to high expressed levels in the sRNA library of G. molesta were all predicted (Fig 3). The 44 miRNAs could be divided into three types: (1) 10 miRNAs derived from either the 5’ or 3’ mature miRNAs of the 5 corresponding precursors, (2) 27 miRNAs only from the 5’ mature miRNAs, and (3) 17 miRNAs only from the 3’ mature miRNAs (Table 3and Fig 3).
Table 3. Novel miRNAs in G. molesta.
| miRNA | Sequence | Length | Seed | CG(%) | Free energy | Reads account |
|---|---|---|---|---|---|---|
| gmo-miR-PC-5p-15_50867 | CGCAATATATTGAATGGGCCTG | 22 | GCAATAT | 48.40 | -33.10 | 127,819 |
| gmo-miR-PC-3p-598_1629 | ACCAAACTTGATCATTTAGAGGAAGT | 26 | CCAAACT | 49.40 | -57.50 | 17,872 |
| gmo-miR-PC-3p-674_1493 | ACCCTGTTGAGCTTGACTCTAGTCT | 25 | CCCTGTT | 50.30 | -53.20 | 17,424 |
| gmo-miR-PC-5p-1258_867 | TACCGATTGAATGATTTAGTGAGGTC | 26 | ACCGATT | 49.40 | -57.50 | 14,307 |
| gmo-miR-PC-5p-1018_1028 | GACCCTGTTGAGCTTGACTCTAGTCT | 26 | ACCCTGT | 50.30 | -53.20 | 9,152 |
| gmo-miR-PC-5p-671_1474 | CGGGAGGAGGAGTGTTAAACT | 21 | GGGAGGA | 65.20 | -89.30 | 1,593 |
| gmo-miR-PC-5p-1276_891 | CCATCAAAGTCGGTTTGTTATA | 22 | CATCAAA | 38.00 | -30.90 | 1,050 |
| gmo-miR-PC-3p-1433_822 | GTGAATCGTATGTAAAAGT | 19 | TGAATCG | 25.20 | -20.20 | 863 |
| gmo-miR-PC-3p-4029_320 | CATCACAGTCGGAGTTCTAGCTT | 23 | ATCACAG | 56.50 | -38.80 | 323 |
| gmo-miR-PC-3p-16268_91 | GCCCCATTCAATATACTGCCGA | 22 | CCCCATT | 48.40 | -33.10 | 164 |
| gmo-miR-PC-5p-35148_41 | TTGTAGAATTGGCGAGAAATTAGAGC | 26 | TGTAGAA | 52.50 | -34.10 | 159 |
| gmo-miR-PC-3p-36514_40 | TGGTGTACCGAACTTTTTGAGT | 22 | GGTGTAC | 37.30 | -31.20 | 121 |
| gmo-miR-PC-3p-20051_73 | TAAGCAATTCTATAGGGTTTG | 21 | AAGCAAT | 28.10 | -26.70 | 102 |
| gmo-miR-PC-5p-94853_15 | TTAGCTGTACATCGGTGGACCT | 22 | TAGCTGT | 43.60 | -55.50 | 97 |
| gmo-miR-PC-5p-130153_12 | ACTCAAAAAGTTCGGTACACC | 21 | CTCAAAA | 38.10 | -54.60 | 72.50 |
| gmo-miR-PC-5p-89338_18 | AATGGGTATGCATCTAGTGGC | 21 | ATGGGTA | 40.50 | -47.60 | 46 |
| gmo-miR-PC-5p-82222_20 | TACTCAAATAGTTCGGTACACC | 22 | ACTCAAA | 37.30 | -31.20 | 39.50 |
| gmo-miR-PC-5p-83411_19 | ATCCTACCGGCTGCGCCA | 18 | TCCTACC | 53.60 | -16.90 | 38 |
| gmo-miR-PC-3p-217431_8 | TGGTGTACCGAACTATTTGACT | 22 | GGTGTAC | 38.10 | -54.60 | 35.50 |
| gmo-miR-PC-5p-66427_22 | TTATACATATTTGTAGAATTGTAGCT | 26 | TATACAT | 30.20 | -41.70 | 35 |
| gmo-miR-PC-5p-193418_8 | TTGGAACAGAGCTGAATTTCATTTGT | 26 | TGGAACA | 29.70 | -30.20 | 26 |
| gmo-miR-PC-5p-93171_18 | ATTAGAATCAGTACGCTTTGTC | 22 | TTAGAAT | 46.40 | -36.40 | 24 |
| gmo-miR-PC-5p-323567_7 | GTCACAAATATCGGAACAACGCCT | 24 | TCACAAA | 45.50 | -66.10 | 20 |
| gmo-miR-PC-3p-803068_3 | NCGTTTTGACGATCGCAAAATG | 22 | CGTTTTG | 44.00 | -33.90 | 20 |
| gmo-miR-PC-5p-113242_14 | TTTAAGTACGTTTTGACCGGCT | 22 | TTAAGTA | 44.90 | -36.60 | 18 |
| gmo-miR-PC-3p-146760_10 | CAGATGGTAAACTCGATACT | 20 | AGATGGT | 40.70 | -35 | 17 |
| gmo-miR-PC-5p-234779_7 | TCACAAATATCTGAACATGCACT | 23 | CACAAAT | 40.70 | -44.20 | 16 |
| gmo-miR-PC-3p-108996_14 | TTCCTTCGTAGACTAAAATGGTAGTT | 26 | TCCTTCG | 28.40 | -19.70 | 16 |
| gmo-miR-PC-5p-237240_7 | GAGGTGGTCATAAATATCGGAACATC | 26 | AGGTGGT | 45.00 | -68.80 | 16 |
| gmo-miR-PC-5p-139696_12 | TCACAAATATCGGAACAACGCCT | 23 | CACAAAT | 40.20 | -28.30 | 15 |
| gmo-miR-PC-5p-272779_7 | AAAAGGGACCTTATTGTCGATGGCGC | 26 | AAAGGGA | 43.90 | -43.30 | 15 |
| gmo-miR-PC-5p-399520_7 | TGGACAAACATTGCTTTACGGGC | 23 | GGACAAA | 52.40 | -22.10 | 13 |
| gmo-miR-PC-5p-390898_5 | TTAGCTGTACATTAGTGGACCT | 22 | TAGCTGT | 44.00 | -58.80 | 13 |
| gmo-miR-PC-3p-180380_9 | ATCCTGTGGAACATCGGATTTGACT | 25 | TCCTGTG | 58.00 | -40.80 | 13 |
| gmo-miR-PC-3p-478770_4 | ACCATGTAGAATTGGAAAACCT | 22 | CCATGTA | 38.50 | -47.30 | 12 |
| gmo-miR-PC-3p-292016_6 | TGACACGCTAGCAACTTCTGCAGC | 24 | GACACGC | 45.80 | -42.30 | 12 |
| gmo-miR-PC-3p-160035_12 | CATGAATATGGTTTTTCCCC | 20 | ATGAATA | 41.60 | -22.70 | 12 |
| gmo-miR-PC-5p-124799_12 | TTGTGATGTGATAGTTGAAGTTGCC | 25 | TGTGATG | 30.90 | -23.50 | 12 |
| gmo-miR-PC-5p-250424_7 | TGACTATATATCGGAAGATGAGCAGG | 26 | GACTATA | 44.00 | -24 | 11 |
| gmo-miR-PC-5p-654402_3 | CAAGATGTCGGCATAGCTGATTT | 23 | AAGATGT | 50.80 | -25.90 | 11 |
| gmo-miR-PC-3p-457779_7 | TTCACTACTGAACAGAGGCCT | 21 | TCACTAC | 54.40 | -40.30 | 11 |
| gmo-miR-PC-5p-150358_10 | TACCGGTATCATTTGTATGTTC | 22 | ACCGGTA | 40.00 | -28.20 | 11 |
| gmo-miR-PC-3p-629556_3 | TAAGGTCCACCAATGTACAGCT | 22 | AAGGTCC | 42.60 | -46.50 | 11 |
| gmo-miR-PC-5p-796447_3 | ATTTCAAATGTCGTAAGTCGCA | 22 | TTTCAAA | 36.70 | -47 | 10 |
gmo: Grapholitha molesta.
Fig 3. Predicted secondary structures of the 44 novel miRNAs with middle to high abundance in the sRNA library of G. molesta.
The mature miRNAs located in the 5’ arm are shaded in yellow, whereas the mature miRNAs located in the 3’ arm are shaded in blue.
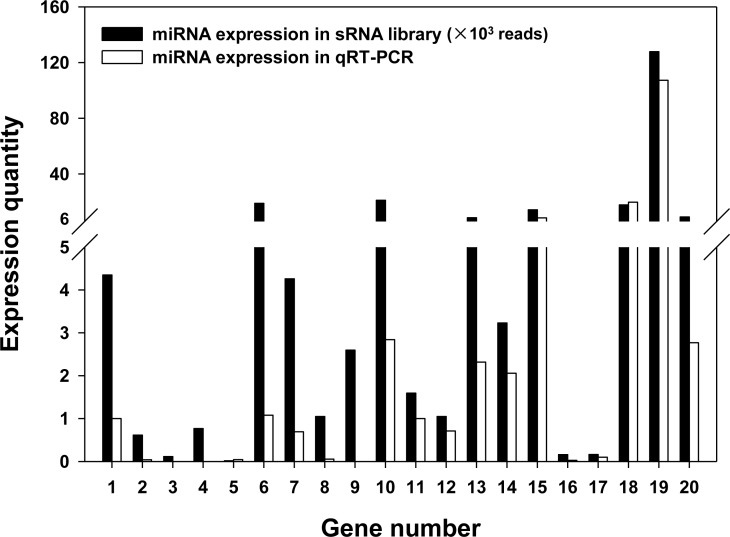
In order to verify the Illumina sequencing, the expressional quantity and sequence accuracy of the 10 conserved and 10 novel miRNA determined in the sRNA library of G. molesta were validated with the qPCR method and Sanger sequencing. The result of qPCR demonstrated that the abundance of most selected miRNAs showed concordant expressional profiles with the corresponding reads accounts in sRNA library, although bmo9a exhibited very low expressional level in the qPCR test (Fig 4). Sequence alignment illustrated that most of the miRNA sequences sequenced by Illumina platform were in accordance with those sequenced by Sanger method. The substitutions at the 5’end produced by the artificially added bases in the forward primer and the differences of one or two bases at the 3’ end resulted from sequencing deviation all have no influence on annotation and target prediction of miRNAs (S7 Table). Therefore, Illumina sequencing is a reliable platform for the overall understanding of miRNA profile in G. molesta.
Fig 4. Comparative analysis of qPCR detection with the expressional profiling in sRNA sequencing.
The y-axis indicates the abundance of miRNAs in the sRNA library and the relative transcriptional level of miRNAs detected using qPCR. The x-axis represents the specific assessed miRNAs, as 1: let-7, 2: mse92a, 3: bmo998, 4: pxy-6497-p5, 5: pxy-6497-p3, 6: bmo2b, 7: bmo281, 8: bmo279a, 9: bmo9a, 10: bmo305, 11: PC-5P-671, 12: PC-5P-1276, 13: PC-3P-1433, 14: PC-3P-4029, 15: PC-3P-16268, 16: PC-5P-35148, 17: PC-5P-15, 18: PC-5P-1018, 19: PC-5P-1258, 20: PC-3P-598.
Cluster of miRNAs
miRNAs that were transcribed from the same primary miRNA (pri-miRNA) frequently exhibit clustered distribution in the genome and were usually expressed as a polycistronic transcript with co-regulation activity in biological networks. The analysis of the miRNA cluster referred to the genome of D. plexippus (Table 4) revealed that more miRNAs in G. molesta were linked within 10–50 kb genomic distances, with 28.92% of miRNAs clustered within 50 kb and 26.51% miRNAs closely linked within 10 kb. There were 22.89% and 21.69% miRNAs respectively clustered within 5 kb and 3 kb genomic distance. The mean number of miRNAs per cluster was 3.2, 2.9, 2.7 and 2.8 correspondingly in 50 kb, 10 kb, 5kb and 3 kb genomic distances. At the same genomic distance, a positive and correlated expression relationship has been normally detected among miRNAs within the same cluster ID, or at least two of which appeared with similar expressional profiles. However, the expression patterns (reads account) between distance-neighboring miRNAs of the same cluster varied among different clusters.
Table 4. miRNA cluster analysis in G. molesta.
| Genomic distance | ||||
|---|---|---|---|---|
| Cluster IDa | 50k | 10k | 5k | 3k |
| 1 | bmo-miR-iab-4-3p_L-3R+1 (10b) | bmo-miR-iab-4-3p_L-3R+1 (10) | bmo-miR-iab-4-3p_L-3R+1 (10) | bmo-miR-iab-4-3p_L-3R+1 (10) |
| bmo-miR-iab-4-5p (59) | bmo-miR-iab-4-5p (59) | bmo-miR-iab-4-5p (59) | bmo-miR-iab-4-5p (59) | |
| bmo-miR-iab-8_R+2 (20) | bmo-miR-iab-8_R+2 (20) | bmo-miR-iab-8_R+2 (20) | bmo-miR-iab-8_R+2 (20) | |
| 2 | bmo-miR-34-5p_R+1 (1573) | bmo-miR-34-5p_R+1 (1573) | gmo-miR-PC-3p-1153688_2 (2) | gmo-miR-PC-3p-1153688_2 (2) |
| bmo-miR-277-3p (8369) | bmo-miR-277-3p (8369) | mse-mir-278-p5 (36) | mse-mir-278-p5 (36) | |
| bmo-miR-277-5p_L-1R+1 (18.5) | bmo-miR-277-5p_L-1R+1 (18.5) | |||
| bmo-miR-317-5p (22) | ||||
| 3 | gmo-miR-PC-3p-1153688_2 (2) | gmo-miR-PC-3p-1153688_2 (2) | gmo-miR-PC-3p-674_1493 (17424) | gmo-miR-PC-3p-674_1493 (17424) |
| mse-mir-278-p5 (36) | mse-mir-278-p5 (36) | gmo-miR-PC-5p-1018_1028 (9152) | gmo-miR-PC-5p-1018_1028 (9152) | |
| bmo-mir-6497-p3 (2985) | bmo-mir-6497-p3 (2985) | |||
| bmo-mir-6497-p5 (24686) | bmo-mir-6497-p5 (24686) | |||
| 4 | gmo-miR-PC-3p-674_1493 (17424) | gmo-miR-PC-3p-674_1493 (17424) | bmo-miR-10-3p (5036) | bmo-miR-10-3p (5036) |
| gmo-miR-PC-5p-1018_1028 (9152) | gmo-miR-PC-5p-1018_1028 (9152) | bmo-miR-10-5p_L+1 (338584) | bmo-miR-10-5p_L+1 (338584) | |
| bmo-mir-6497-p3 (2985) | bmo-mir-6497-p3 (2985) | dme-miR-10-5p_L+1_1ss23TA (3798) | dme-miR-10-5p_L+1_1ss23TA (3798) | |
| bmo-mir-6497-p5 (24686) | bmo-mir-6497-p5 (24686) | |||
| gmo-miR-PC-5p-1258_867 (14307) | gmo-miR-PC-5p-1258_867 (14307) | |||
| 5 | bmo-miR-10-3p (5036) | bmo-miR-10-3p (5036) | bmo-miR-71-3p_L-1R+1 (13455) | gmo-miR-PC-3p-803068_3 (20) |
| bmo-miR-10-5p_L+1 (338584) | bmo-miR-10-5p_L+1 (338584) | gmo-miR-PC-5p-1276_891 (1050) | bmo-miR-274-3p_L+1_1ss10GA (1215) | |
| dme-miR-10-5p_L+1_1ss23TA (3798) | dme-miR-10-5p_L+1_1ss23TA (3798) | pxy-miR-274 (6383) | ||
| pxy-mir-274-p3_1ss10TC (10) | ||||
| 6 | bmo-miR-71-3p_L-1R+1 (13455) | bmo-miR-71-3p_L-1R+1 (13455) | gmo-miR-PC-3p-803068_3 (20) | mse-miR-6094 (19) |
| gmo-miR-PC-5p-1276_891 (1050) | gmo-miR-PC-5p-1276_891 (1050) | bmo-miR-274-3p_L+1_1ss10GA (1215) | mse-mir-6094-p5_1ss7GA (19) | |
| pxy-miR-274 (6383) | gmo-miR-PC-5p-201559_9 (9) | |||
| pxy-mir-274-p3_1ss10TC (10) | ||||
| 7 | gmo-miR-PC-3p-1433_822 (863) | gmo-miR-PC-3p-803068_3 (20) | mse-miR-6094 (19) | dme-miR-8-3p_R+1 (2914) |
| gmo-miR-PC-3p-803068_3 (20) | bmo-miR-274-3p_L+1_1ss10GA (1215) | mse-mir-6094-p5_1ss7GA (19) | dme-miR-8-5p_R+1 (13) | |
| bmo-miR-274-3p_L+1_1ss10GA (1215) | pxy-miR-274 (6383) | gmo-miR-PC-5p-201559_9 (9) | mse-mir-8-p5 (3279) | |
| pxy-miR-274 (6383) | pxy-mir-274-p3_1ss10TC (10) | |||
| pxy-mir-274-p3_1ss10TC (10) | gmo-miR-PC-5p-113242_14 (19) | |||
| gmo-miR-PC-5p-113242_14 (19) | bmo-miR-252-5p (4031) | |||
| bmo-miR-252-5p (4031) | ||||
| 8 | mse-miR-6094 (19) | mse-miR-6094 (19) | dme-miR-8-3p_R+1 (2914) | dme-miR-276a-5p_L-1R-1_1ss22CT (1) |
| mse-mir-6094-p5_1ss7GA (19) | mse-mir-6094-p5_1ss7GA (19) | dme-miR-8-5p_R+1 (13) | dme-miR-276b-3p_R+1_1ss10AC (47) | |
| gmo-miR-PC-5p-201559_9 (9) | gmo-miR-PC-5p-201559_9 (9) | mse-mir-8-p5 (3279) | bmo-miR-276-5p (92) | |
| 9 | dme-miR-8-3p_R+1 (2914) | dme-miR-8-3p_R+1 (2914) | dme-miR-276a-5p_L-1R-1_1ss22CT (1) | bmo-miR-263a-5p_R+4 (14803) |
| dme-miR-8-5p_R+1 (13) | dme-miR-8-5p_R+1 (13) | dme-miR-276b-3p_R+1_1ss10AC (47) | mse-miR-263a_R-1 (21512) | |
| mse-mir-8-p5 (3279) | mse-mir-8-p5 (3279) | bmo-miR-276-5p (92) | ||
| 10 | dme-miR-276a-5p_L-1R-1_1ss22CT (1) | dme-miR-276a-5p_L-1R-1_1ss22CT (1) | bmo-miR-263a-5p_R+4 (14803) | tur-miR-1-3p_L-2R+1 (10) |
| dme-miR-276b-3p_R+1_1ss10AC (47) | dme-miR-276b-3p_R+1_1ss10AC (47) | mse-miR-263a_R-1 (21512) | bmo-miR-1a-5p_R+1 (38) | |
| bmo-miR-276-5p (92) | bmo-miR-276-5p (92) | |||
| 11 | bmo-miR-263a-5p_R+4 (14803) | bmo-miR-263a-5p_R+4 (14803) | tur-miR-1-3p_L-2R+1 (10) | gmo-miR-PC-5p-237240_7 (16) |
| mse-miR-263a_R-1 (21512) | mse-miR-263a_R-1 (21512) | bmo-miR-1a-5p_R+1 (38) | gmo-miR-PC-5p-323567_7 (20) | |
| 12 | bmo-miR-133 (2828) | tur-miR-1-3p_L-2R+1 (10) | gmo-miR-PC-5p-237240_7 (16) | gmo-miR-PC-5p-1071846_2 (5) |
| bmo-mir-133-p5 (3) | bmo-miR-1a-5p_R+1 (38) | gmo-miR-PC-5p-323567_7 (20) | gmo-miR-PC-5p-1000352_3 (4) | |
| tur-miR-1-3p_L-2R+1 (10) | ||||
| bmo-miR-1a-5p_R+1 (38) | ||||
| 13 | gmo-miR-PC-5p-237240_7 (16) | gmo-miR-PC-5p-237240_7 (16) | gmo-miR-PC-5p-1071846_2 (5) | gmo-miR-PC-3p-812930_3 (14.5) |
| gmo-miR-PC-5p-323567_7 (20) | gmo-miR-PC-5p-323567_7 (20) | gmo-miR-PC-5p-1000352_3 (4) | gmo-miR-PC-5p-36514_40 (121) | |
| gmo-miR-PC-5p-130153_12 (72.5) | ||||
| 14 | gmo-miR-PC-5p-1071846_2 (5) | gmo-miR-PC-5p-1071846_2 (5) | gmo-miR-PC-3p-812930_3 (14.5) | |
| gmo-miR-PC-5p-1000352_3 (4) | gmo-miR-PC-5p-1000352_3 (4) | gmo-miR-PC-5p-36514_40 (121) | ||
| gmo-miR-PC-5p-130153_12 (72.5) | ||||
| 15 | gmo-miR-PC-3p-812930_3 (14.5) | gmo-miR-PC-3p-812930_3 (14.5) | ||
| gmo-miR-PC-5p-36514_40 (121) | gmo-miR-PC-5p-36514_40 (121) | |||
| gmo-miR-PC-5p-130153_12 (72.5) | gmo-miR-PC-5p-130153_12 (72.5) | |||
a miRNAs with same cluster ID number belong to the same one pre-miRNA cluster
b The number in the brackets followed the name of miRNA represents the reads account of the corresponding miRNA in the sRNA library of G. molesta.
PC is short for "putative candidate" and represents novel miRNA.
KEGG analysis of predicted target genes
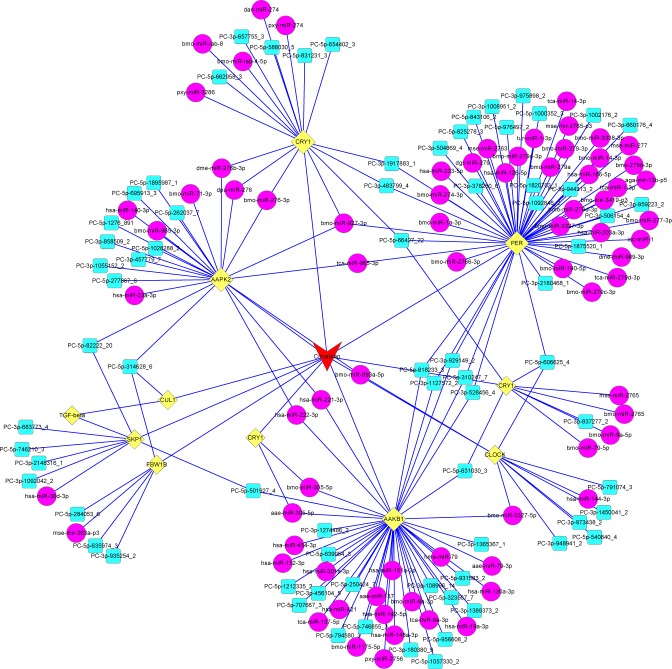
KEGG is a database that helps understanding the molecular interaction and reaction networks in cells and organisms (KEGG PATHWAY Database: http://www.kegg.jp/kegg/pathway.html). In this study, the KEGG analysis of predicted target genes of miRNAs was used to increase our understanding about the biological functions of the identified miRNAs in the metabolism and development of G. molesta. A total of 1,896 target genes of the miRNAs in the G. molesta were significantly enriched in the KEGG analysis, among which 410 genes were matched to 16 KEGG pathways, including 10 pathways of amino acid metabolism and protein processing (ko00270, ko00330, ko00650, ko00250, ko00280, ko00640, ko00350, ko00410, ko04964 and ko04141), 2 pathways involved in nucleotide repairing (ko03420) and mRNA surveillance (ko03015), 1 pathway of plant-pathogen interaction (ko04626), 2 pathways participating in development regulation (ko04350 and ko04391), and 1 pathway involved in circadian rhythm (ko04710) (S8 Table). The further gene network analysis demonstrated that many conserved and novel miRNAs in G. molesta participated in the circadian rhythm through networks among clock genes (cry1, clock and per) and genes involving in growth and development regulation (TGF-beta, skp1, fbw1B, and cul1) (Fig 5).
Fig 5. Network prediction of the circadian rhythm pathway in G. molesta.
The miRNAs in magenta and cyan circles respectively represent the conserved and novel miRNAs. Genes in yellow diamonds are clock genes and those involved in growth and development regulation. The regulatory network of miRNAs and their target genes for the circadian rhythm in G. molesta are linked with blue lines.
Expressional stability of candidate reference genes under different experimental conditions
A universal method, mean expression value normalization (global mean), was adopted for preliminarily assess the expressional stabilities of the candidate reference genes. This method demonstrated with better reduction of technical variation and more accurate appreciation of biological change [34]. After calculation and comparison of the Spearman’s rank correlation coefficient rs, eight candidate reference sRNAs (bmo-miR-2b-3p_1ss22GC, pxy-mir-6497-p3_1ss7CT, hsa-miR-16-5p, bmo-miR-281-3p_L-2R+2, U6 snRNA, bmo-miR-279a_R+2, 5S rRNA and bmo-miR-9a-5p) were preliminarily identified by their positive correlation with the global mean expressions (Table 5), and were further evaluated with geNorm software.
Table 5. Information of candidate reference genes used for qRT-PCR.
| Primer information | Correlation to global mean Ct | ||||||
|---|---|---|---|---|---|---|---|
| Gene name | Symbol | Forward primer sequence(5’-3’)a | Tm (°C) | E (%) | R2 | rsb | pc |
| bmo-miR-2b-3p_1ss22GC | bmo2b | cccATCACAGCCAGCTTTG | 59.7 | 106.5 | 0.995 | 0.952*** | 0 |
| pxy-mir-6497-p3_1ss7CT | pxy-6497-p3 | ccgtGATCTTCCTAGCCGT | 59.7 | 97.3 | 0.995 | 0.857** | 0.007 |
| hsa-miR-16-5p | hsa16 | gaccaggTAGCAGCACGT | 59.6 | 91.4 | 0.978 | 0.833** | 0.01 |
| bmo-miR-281-3p_L-2R+2 | bmo281 | gcggcTGTCATGGAGTTG | 59.6 | 102.8 | 0.993 | 0.833** | 0.01 |
| U6 snRNA | U6 | CGCAAGGATGACACGCAA | 57.3 | 126.8 | 0.991 | 0.833** | 0.01 |
| bmo-miR-279a_R+2 | bmo279a | ccgtccgGATCCACACTC | 61.9 | 97.4 | 0.956 | 0.69 | 0.058 |
| 5S rRNA | 5S | GCAGTCCACCGAAGTTAAGC | 59.8 | 85.8 | 0.997 | 0.407 | 0.317 |
| bmo-miR-9a-5p | bmo9a | gtcggtcgGTTATCTAGCT | 57.6 | 94.1 | 0.998 | 0.347 | 0.399 |
| bmo-miR-998_R+2 | bmo998 | gtcagagaGCACCATGGGA | 59.7 | 91.4 | 0.998 | -0.048 | 0.911 |
| bmo-miR-305-5p_R+1 | bmo305 | ggcagtGTACTTCATCAGG | 57.6 | 90.7 | 0.998 | -0.071 | 0.867 |
| bmo-let-7-5p | let-7 | cgcgcTGAGGTAGTAGGTTG | 61.9 | 84.6 | 0.979 | -0.143 | 0.736 |
| mse-miR-92a | mse92a | ccatcccGCACCAGTCC | 61.8 | 102.3 | 0.985 | -0.429 | 0.289 |
a The upper case letters are nucleotides from corresponding miRNA, and the lower case letters are artificially added for adjustment of GC content in the primer
b Spearman’s rank correlation coefficient
c Two-tailed tests
*** represents P≤0.001
** represents 0.001<P≤0.01.
geNorm software ranks the reference gene according to their M values (average expression stability) and provides optimal recommendation of the number of reference genes based on the pairwise variation (V) analysis. In this study, the expressional stabilities of the 8 candidate reference genes were ranked under eight different experimental conditions (S2 Fig). The candidate reference, bmo2b, was ranked with high stability under seven experimental conditions, except for temperature treatment; U6 and bmo9a were all ranked as stable references under four experimental conditions. In combination with the V value (S3 Fig), the recommendations of internal normalizers for analysis of miRNA expressional profiles of G. molesta using qRT-PCR were determined for each of the eight specific experimental conditions. For intrinsic biotic conditions, 3 sRNAs (hsa16, U6 and bmo2b) used together were recommended for study of different developmental stages. Combined usage of 6 sRNAs (hsa16, bmo2b, U6, 5S, bmo279 and bmo9a) was recommended for miRNA quantification in different tissues (Table 6). For different exogenous stresses, it is recommended 2 sRNAs (bmo281 and U6) for temperature changes, 3 miRNAs (bmo2b, bmo279 and hsa16) for photoperiod treatment, 2 miRNAs (bmo279 and bmo9a) for starvation, 2 miRNAs (bmo2b and bmo9a) for JH exposure, 2 miRNAs (bmo2b and bmo281) for dsRNA treatment, and 2 sRNAs (bmo281 and U6) for insecticide application (Table 6).
Table 6. Recommended reference genes for expressional qualification of microRNA in G. molesta under different experimental conditions.
| Experimental conditions | Recommended reference genes | |||||
|---|---|---|---|---|---|---|
| Intrinsic biotic factors | ||||||
| Developmental stage | hsa16 | U6 | bmo2b | |||
| Tissue | hsa16 | bmo2b | U6 | 5S | bmo279a | bmo9a |
| Exogenous impact factors | ||||||
| Temperature | bmo281 | U6 | ||||
| Photoperiod | bmo2b | bmo279a | hsa16 | |||
| Starvation | bmo279a | bmo9a | ||||
| JH injection | bmo2b | bmo9a | ||||
| dsRNA injection | bmo2b | bmo281 | ||||
| Insecticide | bmo281 | U6 | ||||
Validation of reference gene selection
To evaluate the performance of selected reference genes, the expression level of let-7, which participates in development regulation in insects [58,59] and showed dynamic expressional profiles in different developmental stages of Bombyx mori [60] was examined in different developmental stages of G. molesta. Similar expression profiles of let-7 in different developmental stages of G. molesta were detected when single best recommended reference NF(1) (hsa16) and the optimal recommendation NF(1–3) (hsa16 + U6 + bmo2b) were used as normalization factors, only with different expression patterns in the second vs third instars and fifth instar vs male pupa (Table 7). However, the use of single least stable reference NF(8) and NF(12), respectively, showed significant down-regulation and up-regulation of let-7 within all tested developmental stages of G. molesta (Table 7).
Table 7. Expressional profiles of let-7 in G. molesta quantified according to different normalizers.
| Normalizers§ | ||||
|---|---|---|---|---|
| Developmental stage | NF(1) | NF(1–3) | NF(8) | NF(12) |
| Egg | 1±0.06a# | 1±0.04a | 1±0.03a | 1±0.06a |
| 1st instar | 0.34±0.02c | 0.77±0.02b | 0d | 3.77±0.09a |
| 2nd instar | 28.86±5.81c | 63.84±3.88b | 0.62±0.03c | 292.08±19.09a |
| 3rd instar | 39.29±3.44b | 53.36±1.59b | 0.24±0.01c | 382.84±28.82a |
| 4th instar | 23.74±0.82c | 49.83±0.8b | 0.22±0.02d | 284.74±10.86a |
| 5th instar | 93.29±13.37b | 150.21±13.85a | 0.53±0.07c | 78.45±8.25b |
| Prepupa | 50.8±3.33a | 54.36±1.01a | 0.22±0.02c | 19.86±0.17b |
| Male-pupa | 259.19±26.86a | 141.35±4.93b | 6.61±0.16c | 65.62±1.79c |
| Female-pupa | 458.93±25.01a | 338.23±7.01b | 11.06±0.75d | 93.34±4.92c |
| Male-adult | 503.95±19.2b | 413.02±29.07b | 0.96±0.07c | 3325.75±272.5a |
| Female-adult | 120.02±7.43b | 106.65±7.58b | 0.93±0.04c | 239.26±10.66a |
§ Different normalization factors: NF (1), using the single one best reference hsa16 for normalization; NF(1–3), using the recommended combination of the three best reference sRNAs, hsa16, U6 and bmo2b for normalization; NF (8), using the single least stable reference bmo281 for normalization; NF(12), using single mse92a with the lowest rs value for normalization
# Means within the same line followed by the different letters are significantly different (P ≤ 0.05).
Discussion
The miRNA information of several Lepidopteran insects has been identified with reference to their genomic information [20,61] or genome sequences of the model insect silkworm (B. mori) [62,63]. In the present study, a pooled sRNA library of G. molesta was prepared from mixed developmental stages of oriental fruit moth, and was further analyzed in reference to its own transcriptome and the genome of D. plexippus which was found with the highest similarity to the transcripts of G. molesta in the phylogenetic conservation analysis (unpublished data). A total of 536 mature miRNAs composed of 291 previously reported and 245 novel ones were finally identified in G. molesta. In previous studies of Lepidopteran insects, 55 conserved and 202 novel miRNAs were found in B. mori [61], 163 conserved and 13 novel miRNAs were identified in M. sexta [20], and 97, 91 and 69 conserved together with 1, 8 and 383 novel ones were respectively identified in the miRNA analyses of Helicoverpa armigera, Spodoptera litura and P. xylostella [62,63]. With the cumulative availability of miRNA data and the increase of sequencing depth, more information of mature miRNA would be uncovered and more novel miRNA could be identified in lepidopterans, which would provide useful repertoires for clarification of the modulation complex in insects and other organisms.
Among the 291 conserved miRNAs identified in the sRNA transcriptome of G. molesta, 40 miRNAs showed highly expressed levels, which probably play critical roles in various regulatory processes in the oriental fruit moth. The miRNAs with the highest account of reads in this study including miR-10, miR-8, miR-281, and miR-263 were also identified as the most abundantly expressed ones in A. gambiae [64]. miR-10 and miR-8 were found with high abundance in the digestive tract of green bottle fly maggot Lucilia sericata and both were considered involved in secretions [65]. A study on S. exigua demonstrated that miR-10 also participated in developmental regulation since oral feeding using synthetic miRNA mimics of miR-10-1a resulted in suppressed growth and increased mortality [66]. miR-8 showed pleiotropic regulatory functions in insects, including neurogenesis through targeting atrophin mRNA or regulating synaptic activity [67,68], homeostasis of innate immunity by altering the expression of antimicrobial peptides [69], and development tuning via responding to ecdysone [70]. miR-281 is another multi-functional miRNA in insects. It is involved in regulation of long chain fatty acid synthesis and cuticle formation in M. sexta [71], highly expressed in Malpighian tubules and participating in developmental regulation through targeting and suppressing the transcription of ecdysone receptor–B (EcRB) in B. mori [72], and facilitating the replication of dengue virus (DENV-2) in vector mosquito Aedes albopictus by specifically induced in female midgut upon DENV-2 infection [73]. The conserved miR-263 was abundantly expressed in pupae and may participate in temporal regulation during silkworm development [74]. The other two miRNAs, miR-276 and miR-14 that showed reads account at top levels in the sRNA library of G. molesta, were also found with regulatory effects in the development, reproduction and immunity of insects. miR-276 was noted as one of the miRNAs for identifying reproductive stages of A. mellifera independent of caste [75] and was found promoting egg-hatching synchrony of locusts through up-regulation of the target, a transcription coactivator gene brahma (brm) [10]. In studies of Drosophila, miR-14 has been considered as an important regulator in the development process, mainly through negative modulation of the Hedgehog signaling [76], positive regulation of the autoregulatory loop of ecdysone signaling [77] and autophagy regulation during salivary gland cell death by targeting 1,4,5-triphospate (IP3) signaling [78]. miR-14 was also found involved in the interactions of insect and virus, e.g. H. armigera single nucleopolyhedrovirus (HaSNPV) manipulated EcR transcription in H. armigera through down regulation of miR-14 in host insect [79] and miR-14 in the white-backed planthopper Sogatella furcifera may participate in the immune response against the southern rice black-streaked dwarf virus vectored by the planthopper [80].
Analysis about organisms of different kingdoms demonstrated that large fractions of miRNAs share conservation among closely-related species and even species with distant phylogenetic relationships, suggesting evolutionarily conserved regulatory functions across species [81]. In the present study, the 79 miRNA families found in the sRNA library of G. molesta were detected to be conserved among the Lepidoptera insects, yet most of the miRNA families numbered from miR-1175 to miR-8506 were absent in other species. More miRNAs among the 79 families were detected to be present in Hexapoda species, whereas notably less number of miRNA families were found in species in Brachiopoda, Chiopoda and Arachinida, although they all belong to Arthropods. Let-7 and miR-124 were conserved among Arthropoda, Nematoda and Chordata species in our analysis. Previous analyses suggested that many miRNA families, such as let-7, miR-10, miR-99 and miR-125 were highly conserved among animal species [46], whereas miR-99 was not found in the sRNA library of G. molesta, while miR-10 and miR-125 were conserved in most species but absent in C. elegans. Numerous studies have demonstrated that miRNAs showed widely conservative evolution, and thus have been proposed to be used as additional sequence markers in combination of mitochondrial or gnomic DNA for evolutionary modeling and phylogenetic analysis among various taxa [82,83]. Insects have been considered as an excellent model for study about regulatory functions of miRNAs in conserved regulatory pathways between vertebrates and invertebrates [3]. Besides of sequence conservation, miRNAs were also demonstrated with spatial and temporal expression conservation. A comparative study among bilaterian animals suggested that the tissue specificity of ancient microRNAs was highly conserved [84]. A recent study on Drosophila discovered that the temporal expression of orthologous microRNAs could be more conserved than their sequences and the hourglass pattern of miRNA expression are highly similar, not only among the miRNAs with highly conserved sequences, but also for rapidly evolving orthologs [85]. Therefore, as an important regulation component at posttranscriptional level between mRNA and gene-encoding protein, miRNA and its expression pattern were considered critical in understanding the evolution of developmental gene expression.
Similar to previous studies in planthopper L. striatellus [46] and diamondback moth P. xylostella [63], the abundances of novel miRNAs were also found very low in G. molesta and the 5 novel miRNAs with the highest abundance may play important roles in the development of G. molesta. The secondary structure prediction demonstrated that only a small fraction of mature ones were generated from both arms and most of the mature novel miRNAs were produced from either the 3’ or 5’ arms of the hairpins with a slight bias toward 5’ arm usage (5p/3p proportion was around 1.59). The arm usage preference differed in various Diptera, such as 5’ arm-bias in Drosophila [86], but 3’ arm-bias in A. gambiae [64]. Studies in mammalians illustrated that selection and usage of preferred arm could be dynamically regulated in a development and tissue specific pattern [87]. The study in A. gambiae revealed that arm-switching event could happen after blood feeding [64], all of which suggest that miRNAs are expressed under a condition-specific manner and arm switching can significantly enrich the regulatory capacity of miRNA.
As the next-generation sequencing (NGS) method, Illumina sequencing enables the high-throughput and comprehensive understanding about the genome, transcriptome, epigenome or microbiome of a variety of organisms [88–91]. However, Illumina sequencing has been found more prone to produce deviation than Sanger technique, as it is vulnerable to be contaminated in the process of library preparation and accompanied with deviations resulted from base bias and subjective supposition in the bio-information analysis. In the present study, qPCR and Sanger sequencing were adopted for validation of the sRNA library of G. molesta sequenced by Illumina platform and concordant results between different methods were detected for most of the miRNAs. The divergences between Illumina sequencing and qPCR were mainly due to the limited replication of Illumina analysis, thus, qPCR validation is indispensible for sophisticated measurement about the expressional pattern of specific miRNA of interest. Alignment detected sequence differences happened in some miRNAs between Sanger and Illumina sequencing results. However, most parts of these miRNA sequences were in agreement between the two methods and the seed regions for target prediction were intact after eliminating the sequence deviations at the 5’ end caused by primer modification for adjustment of GC content. Therefore, Illumina sequencing is a reliable platform for the overall understanding of miRNA profile in G. molesta.
Cluster analysis is an important part for understanding the biological network and evolutionary conservation of miRNA. In this study, the clusters of the conserved and novel miRNAs determined in G. molesta were analyzed in reference to the genome of D. plexippus. Fifteen clusters were identified separately at 50 kb, 10 kb, 5kb and 3 kb genomic distances. Most of the miRNAs in the same cluster showed concordant expressional abundance within the same genomic distance. A study showed that expressional levels of clustered miRNAs in mosquito libraries were highly correlated and the mature miRNAs in the same cluster exhibited a strong bias towards the same arm selection [64]. However, the arm selection of the miRNAs in the same cluster showed in a more dynamic manner in this study, with only a small bias towards the 5’ arm selection in the same cluster. Conservation of miRNA clusters among related species would provide evolutionary evidence for functional shift of miRNA in insects. A study found that the cluster organized in mir-2/mir-13/mir-71 was highly conserved in insects [86], which was not detected in G. molesta. Cluster analysis is highly dependent upon the corresponding genome sequences, hence in future, with the information of genome sequence of G. molesta and the availability of sequence information in its phylogenetic related species, more useful information of miRNA data would probably be revealed through comparative studies among agricultural pests.
Target prediction and KEGG pathway analysis are important to understand the regulatory networks of miRNAs. In this study, except for the majority pathways in the processing, repairing and surveillance of nucleic acid, amino acid and protein plus one pathway function in the plant-pathogen interaction, three pathways for controlling the growth and development modulation (including two pathways involved in development regulation and one pathway participating in circadian rhythm) were identified in the sRNA library constructed from different developmental stages of G. molesta. The network prediction of the identified circadian rhythm pathway further revealed the close relationship among a range of miRNAs, clock genes and developmental regulation genes in G. molesta. A majority of insect developmental processes exhibit circadian rhythm (including growth, ecdysis, metamorphosis, diapause, and eclosion) and developmental circadian rhythm can be profoundly impacted by environmental changes, especially the daily or seasonally thermal and photoperiodic alterations [92]. Correspondingly, most organisms have evolved circadian clock to adjust their development and metamorphism rhythm for coping with environmental changes [93]. Recent studies have detected that the miRNA might be an important regulator of circadian rhythmicity at posttranscriptional level in various taxa of organisms [94,95]. The overall understanding about the network of miRNA and their target genes in the developmental regulation of insects would provide useful information for the monitoring and management of agricultural pest.
Since the discovery about the vital roles of miRNA at post-transcriptional level, precisely expressional profiles of miRNA under specific conditions have been considered as the prerequisite for understanding its regulatory functions. Identical to the quantification of coding genes, using suitable normalizers synchronically processed with tested genes is also considered as an effective way for eliminating the errors from materials and experimental manipulation and realizing accurate evaluation of miRNA profiles [96]. In recent years, numerous studies on selection and validation of reference genes for miRNA quantification have been published being relevant to plants, human, vertebrates and nematode [45,47,97,98], whereas few studies have been reported about insects. In the present study, on the basis of the sRNA library of G. molesta, one small nuclear RNA U6, one small ribosomal RNA 5SrRNA and ten miRNAs were preliminarily selected as candidate references according to the previous studies [45–47,51–54] and their stabilities under two biotic and six abiotic conditions were further assessed with global mean method and geNorm algorithm. Normally, four statistical methods, geNorm, Normfinder, delta Ct and Bestkeeper are adopted for expressional stability analysis. Based on different algorithms, however, inconsistent outcomes are usually obtained from the four software. Therefore, the comprehensive ranking of expressional stability is hard to determine from parallel analysis with the four algorithms. In the present study, “global mean”, a universal method for analyzing the transcriptional stability of microRNA [34], was adopted for preliminary screening of the candidate reference genes. A final recommendation of suitable normalizers was then clearly acquired with further evaluation by geNorm. In the assessment of reliable internal controls for miRNA expressional quantification in C. elegans, a genome-wide RNA-seq was conducted for preliminary selection of control miRNAs with minimal variation, and similarly, the final stabilities of candidate reference miRNAs were then ranked by applying the common geNorm logarithm [97]. The comprehensive analysis in the present study detected that intrinsic biotic factors including developmental stages and tissues impacted more on the stability of miRNA expression in G. molesta, as a greater number of reference genes should be used for normalization in samples of development stages or tissues. Similarly, investigation in the Chinese perch revealed that embryonic developmental stage was an important factor to the variability of miRNA expression [47]. In previous studies of miRNA evaluation, U6 has always been empirically chosen as an internal control and U6 has been recommended as a stable reference gene under half of the tested conditions in our study. However, the expressional stabilities of U6 and other small nuclear RNAs were actually not be acceptable as endogenous controls in many experimental conditions [45,47]. The present and previous studies all illustrated that misusage of an internal control gene would lead to notably distorted results and misunderstanding of gene expressional profiles [99].
In summary, a total of 536 mature miRNAs including 291 conserved and 245 novel miRNAs was identified in G. molesta. The abundance, conservation and cluster of identified miRNA were analyzed. The KEGG pathway analysis and network prediction of target genes demonstrated that the network composed of miRNAs, clock genes and developmental regulation genes probably play critical roles in the regulation of developmental circadian in G. molesta. Furthermore, suitable reference genes were selected and validated for study on miRNA expressional profile in G. molesta under two biotic and six abiotic experimental conditions. The present study provides an overview of miRNA profile in G. molesta and may serve as a basic reference for evaluation of miRNA abundance in this pest and other insects. Further studies, such as analyzing of the interaction relationship between miRNAs and their targets and deciphering of the regulatory functions and mechanisms of specific miRNAs will shed light on the deeper interpretation of the miRNA-involved post-transcriptional regulation in G. molesta and might also provide a useful foundation for development of new targets or genetic-based techniques for agricultural pest control in the future.
Supporting information
34.7% was mapped to miRBase, 38.06% was mapped to the transcriptome of G. molesta and 27.24% were mapped to the genome of D. plexippus.
(TIF)
The average expression stability (M value) was calculated for each candidate and the sRNA with the lowest M value is considered as the most stably expressed reference gene.
(TIF)
Average pairwise variations (V values) were calculated between the normalization factors NFn and NFn+1, and the addition of reference gene is not required when the V value is below 0.15.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The authors thank Dr. Huaijiang Zhang (Institute of Pomology in Liaoning province) for the sample collection of G. molesta. Special thanks go to Prof. Qingxiao Li (University of Hawaii at Manoa) for copyediting our manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by Chinese Universities Scientific Fund (No. 2014XJ018) and Modern Agro-industry Technology Research System Grants (No. CARS-29-08). The funders had no role in study design, data collection and analysis, decision to publish, and preparation of the manuscript.
References
- 1.Bartel DP. MicroRNA: genomics, biogenesis, mechanism, and function. Cell. 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 2.Cullen BR. Virus and microRNAs. Nat Genet. 2006; 38: S25–30. 10.1038/ng1793 [DOI] [PubMed] [Google Scholar]
- 3.Asgari S. MicroRNA functions in insects. Insect Biochem Mol Biol. 2013; 43: 388–397. 10.1016/j.ibmb.2012.10.005 [DOI] [PubMed] [Google Scholar]
- 4.Lucas K, Raikhel A. Insect microRNAs: Biogenesis, expression profiling and biological functions. Insect Biochem Mol Biol. 2013; 43: 24–38. 10.1016/j.ibmb.2012.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miska EA. How microRNA control cell division, differentiation and death. Curr Opin Genet Dev. 2005; 15: 563–568. 10.1016/j.gde.2005.08.005 [DOI] [PubMed] [Google Scholar]
- 6.Hussain M, Asgari S. Functional analysis of a cellular microRNA in insect host-ascovirus interaction. J Virol. 2010; 84: 612–620. 10.1128/JVI.01794-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chawla G, Sokol NS. MicroRNAs in Drosophila development. Int Rev Cell Mol Biol. 2011; 286: 1–65. 10.1016/B978-0-12-385859-7.00001-X [DOI] [PubMed] [Google Scholar]
- 8.Zdanowicz A, Thermann R, Kowalska J, Jemielity J, Duncan K, Preiss T, et al. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol Cell. 2009; 35: 881–888. 10.1016/j.molcel.2009.09.009 [DOI] [PubMed] [Google Scholar]
- 9.Hussain M, Torres S, Schnettler E, Funk A, Grundhoff A, Pijlman GP, et al. West Nile virus encodes a microRNA-like small RNA in the 3’ untranslated region which unregulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. 2012; 40: 2210–2223. 10.1093/nar/gkr848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He J, Chen Q, Wei Y, Jiang F, Yang M, Hao S, et al. MicroRNA-276 promotes egg-hatching synchrony by up-regulating brim in locusts. Proc Natl Acad Sci U S A. 2016; 113: 584–589. 10.1073/pnas.1521098113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complimentarity to lin-14. Cell. 1993; 75: 843–854. [DOI] [PubMed] [Google Scholar]
- 12.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011; 39: D152–D157. 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Maines JZ, Tastan OY, McKearin DM, Buszczak M. Mei-P26 regulates the maintenance of ovarian germline stem cells by promoting BMP signaling. Development. 2012; 139: 1547–1556. 10.1242/dev.077412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becam I, Rafel N, Hong X, Cohen SM, Milán M. Notch-mediated repression of bantam miRNA contributes to boundary formation in the Drosophila wing. Development. 2011; 138: 3781–3789. 10.1242/dev.064774 [DOI] [PubMed] [Google Scholar]
- 15.Surridge AK, Lopez-Gomollon S, Moxon S, Maroja LS, Rathjen T, Nadeau NJ, et al. Characterisation and expression of microRNAs in developing wings of the neotropical butterfly Heliconius melpomene. BMC Genomics. 2011; 12: 62 10.1186/1471-2164-12-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun K, Westholm JO, Tsurudome K, Hagen JW, Lu Y, Kohwi M, et al. Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants. PLoS Genet. 2012; 8: e1002515 10.1371/journal.pgen.1002515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu P, Gue M, Hay BA. MicroRNA and the regulation of cell death. Trends Genet. 2004; 20: 617–624. 10.1016/j.tig.2004.09.010 [DOI] [PubMed] [Google Scholar]
- 18.Legeai F, Rizk G, Walsh T, Edwards O, Gordon K, Lavenier D, et al. Bioinformatic prediction, deep sequencing of microRNAs and expression analysis during phenotypic plasticity in the pea aphid, Acrythosiphon pisum. BMC Genomics. 2010; 11: 281 10.1186/1471-2164-11-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu F, Peng W, Li Z, Li W, Li L, Pan J, et al. Next-generation small RNA sequencing for microRNAs profiling in Apis mellifera: comparison between nurses and foragers. Insect Mol Biol. 2012; 21: 297–303. 10.1111/j.1365-2583.2012.01135.x [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Zheng Y, Jagadeeswaran G, Ren R, Sunkar R, Jiang H. Identification and development profiling of conserved and novel microRNAs in Manduca sexta. Insect Biochem Mol Biol. 2012; 42: 381–395. 10.1016/j.ibmb.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cayirlioglu P, Kadow IG, Zhan X, Okamura K, Suh GS, Gunning D, et al. Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science. 2008; 319: 1256–1260. 10.1126/science.1149483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenberg JK, Xia J, Zhou X, Thatcher SR, Gu X, Ament SA, et al. Behavioral plasticity in honey bees in associated with differences in brain microRNA transcriptome. Genes brain Behav. 2012; 11: 660–670. 10.1111/j.1601-183X.2012.00782.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009; 137: 273–283. 10.1016/j.cell.2009.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008; 454: 780–783. 10.1038/nature07103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain M, Frentiu FD, Moreira LA, O’Neill SL, Asgari S. Wolbachia utilizes host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc Natl Acad Sci U S A. 2011; 108:9250–9255. 10.1073/pnas.1105469108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci U S A. 2005; 102: 18017–18022. 10.1073/pnas.0508823102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S, Gao S, Zhang D, Yin J, Xiang Z, Xia Q. MicroRNAs show diverse and dynamic expression patterns in multiple tissues of Bombyx mori. BMC Genomics. 2010; 11: 85 10.1186/1471-2164-11-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol. 2003; 259: 9–18. [DOI] [PubMed] [Google Scholar]
- 29.Yu X, Zhou Q, Li S, Luo Q, Cai Y, Lin W, et al. The silkworm (Bombyx mori) microRNAs and their expressions in multiple developmental stages. PLoS ONE. 2008; 3: e2997 10.1371/journal.pone.0002997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson PT, Baldwin DA, Scearce LM, Oberhotzer JC, Tobias JW, Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat Methods. 2004; 1: 155–161. 10.1038/nmeth717 [DOI] [PubMed] [Google Scholar]
- 31.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005; 435: 834–838. 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 32.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005; 33: e179 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan G, Lim QE, Too HP. High-performance quantification of mature microRNAs by real-time RT-PCR using deoxyuridine-incorporated oligonucleotides and hemi-nested primers. RNA. 2010; 16: 1436–1445. 10.1261/rna.2001610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009; 10: R64 10.1186/gb-2009-10-6-r64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Q, Li Z, Cao J, Zhang S, Zhang H, Wu X, et al. Selection and assessment of reference genes for quantitative PCR normalization in migratoty locust Locusta migratoria (Orthoptera: Acrididae). PLoS ONE. 2014; 9: e98164 10.1371/journal.pone.0098164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, An S, Li Z, Wu F, Yang Q, Liu Y, et al. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera. Gene. 2015; 555: 393–402. 10.1016/j.gene.2014.11.038 [DOI] [PubMed] [Google Scholar]
- 37.Kirk H, Dorn S, Mazzi D. Worldwide population genetic structure of the oriental fruit moth (Grapholita molesta), a globally invasive pest. BMC Ecol. 2013; 13: 12 10.1186/1472-6785-13-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothschild GHL, Vickers RA. Biology, ecology and control of the oriental fruit moth In: vander Geest LPS, Evenhuis HH, editors. World crops pest, Tortricid pests their biology, natural enemies and control. Amsterdan: Elsevier Academic Publishers; 1991. pp. 389–412. [Google Scholar]
- 39.Li G, Zhang Y, Li Y, Wu J, Xu X. Cloning, expression, and functional analysis of three odorant-binding proteins of the oriental fruit moth, Grapholita molesta (Busck) (Lepidoptera: Tortricidae). Arch Insect Biochem Physiol. 2016; 91: 67–87. 10.1002/arch.21309 [DOI] [PubMed] [Google Scholar]
- 40.Mazzi D, Dorn S. Movement of insect pests in agricultural landscapes. Ann Appl Biol. 2012; 160: 97–113. [Google Scholar]
- 41.Myers CT, Hull LA, Krawczyk G. Effects of orchard host plants (apple and peach) on development of oriental fruit moth (Lepidoptera: Tortricidae). J Econ Entomol. 2007; 100: 421–430. [DOI] [PubMed] [Google Scholar]
- 42.Kanga LHB, Pree DJ, van Lier JL, Walker GM. Management of insecticide resistance in oriental fruit moth (Grapholita molesta; Lepidoptera: Tortricidae) populations from Ontario. Pest Manag Sci. 2003; 59: 921–927. 10.1002/ps.702 [DOI] [PubMed] [Google Scholar]
- 43.Tollerup KE, Rucker A, Shearer PW. Whole-farm mating disruption to manage Grapholita molesta (Lepidoptera: Tortricidae) in diversified New Jersey Orchards. J Econ Entomol. 2012; 105: 1712–1718. [DOI] [PubMed] [Google Scholar]
- 44.Cao J, Liu Y, Yang Y, Zhang H, Li Z, Yang Q, et al. Molecular characterization and functional analysis of the ultraspiracle (USP) in the oriental fruit moth Grapholita molesta (Lepidoptera: Olethreutidae). Comp Biochem physiol B Biochem Mol Biol. 2015; 190: 54–62. 10.1016/j.cbpb.2015.08.008 [DOI] [PubMed] [Google Scholar]
- 45.Torres A, Torres K, Wdowiak P, Paszkowski T, Maciejewski R. Selection and validation of endogenous controls for microRNA expression studies in endometrioid endometrial cancer tissues. Gynecol Oncol. 2013; 130: 588–594. 10.1016/j.ygyno.2013.06.026 [DOI] [PubMed] [Google Scholar]
- 46.Mahdipour M, van Tol HTA, Stout TAE, Roelen BAJ. Validating reference microRNAs for normalizing qRT-PCR data in bovine oocytes and preimplantation embryos. BMC Dev Biol. 2015; 15: 25 10.1186/s12861-015-0075-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu X, Li Y, Chen D, Wu P, Yi T, Chen T, et al. Selection of reference genes for microRNA quantitative expression analysis in Chinese perch, Siniperca chuatsi. Int J Mol Sci. 2015; 16: 8310–8323. 10.3390/ijms16048310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wheat CW, Wahlberg N. Phylogenomic insights into the cambrian explosion, the colonization of land and the evolution of flight in arthropoda. Syst Biol. 2013; 62: 93–109. 10.1093/sysbio/sys074 [DOI] [PubMed] [Google Scholar]
- 49.Zhou G, Wang T, Lou Y, Cheng J, Zhang H, Xu J. Identification and characterization of microRNAs in small brown planthopper (Laodephax striatellus) by next-generation sequencing. PLoS ONE. 2014; 9: e103041 10.1371/journal.pone.0103041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucl Acids Res. 2008; 36: D480–484. 10.1093/nar/gkm882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gharbi S, Shamsara M, Khateri S, Soroush MR, Ghorbanmehr N, Tavallaei M, et al. Identification of reliable reference genes for quantification of microRNAs in serum samples of sulfur mustard-exposed veterans. Cell J. 2015; 17: 494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han H, Jo YN, Lee JY, Choi SY, Jeong Y, Yun J, et al. Identification of suitable reference genes for the relative quantification of microRNAs in pleural effusion. Oncol Lett. 2014; 8: 1889–1895. 10.3892/ol.2014.2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008; 14: 844–852. 10.1261/rna.939908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin Y, Lai Z. Evaluation of suitable reference genes for normalization of microRNA expression by real-time reverse transcription PCR analysis during longan somatic embryogenesis. Plant physiol Biochem. 2013; 66: 20–25. 10.1016/j.plaphy.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 55.Pfaffl MW, Tichopad A, Progomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: Bestkeeper-Excel-based tool using pair-wise correlations. Biotechnol let. 2004; 26: 509–515. [DOI] [PubMed] [Google Scholar]
- 56.Biosen MK, Dehlendorff C, Linnemann D, Schultz NA, Jensen BV, Høgdall EVS, et al. MicroRNA expression in formalin-fixed paraffin-embedded cancer tissue: identifying reference microRNAs and variability. BMC Cancer. 2015; 15: 1024 10.1186/s12885-015-2030-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biol. 2002; 3: research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rubio M, Belles X. Subtle roles of microRNAs let-7, miR-100 and miR-125 on wing morphogenesis in hemimetabolan metamorphosis. J Insect Physiol. 2013; 59: 1089–1094. 10.1016/j.jinsphys.2013.09.003 [DOI] [PubMed] [Google Scholar]
- 59.Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev Biol. 2002; 244: 170–179. 10.1006/dbio.2002.0594 [DOI] [PubMed] [Google Scholar]
- 60.Liu S, Xia Q, Zhao P, Cheng T, Hong K, Xiang Z. Characterization and expression patterns of let-7 microRNA in the silkworm (Bombyx mori). BMC Dev Biol. 2007; 7: 88 10.1186/1471-213X-7-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu S, Li D, Li Q, Zhao P, Xiang Z, Xia Q. MicroRNAs of Bombyx mori identified by solexa sequencing. BMC Genomics. 2010; 11: 148 10.1186/1471-2164-11-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ge X, Zhang Y, Jiang J, Zhong Y, Yang X, Li Z, et al. Identification of microRNAs in Helicoverpa armigera and Spodoptera litura based on deep sequencing and homology analysis. Int J Biol Sci. 2013; 9: 1–15. 10.7150/ijbs.5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liang P, Feng B, Zhou X, Gao X. Identification and developmental profiling of microRNAs in diamondback moth, Plutella xylostella (L.). PLoS ONE. 2013; 8: e78787 10.1371/journal.pone.0078787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Biryukova I, Ye T, Levashina E. Transcriptome-wide analysis of microRNA expression in the malaria mosquito Anopheles gambiae. BMC Genomics. 2014; 15: 557 10.1186/1471-2164-15-557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blenkiron C, Tsai P, Brown LA, Tintinger V, Askelund KJ, Windsor JA, et al. Characterisation of the small RNAs in the biomedically important green-bottle blowfly Lucilia sericata. PLoS ONE. 2015; 10: e0122203 10.1371/journal.pone.0122203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Huang Q, Yin G, Lee S, Jia R, Liu Z, et al. Identification of microRNAs by small RNA deep sequencing for synthetic microRNAs mimics to control Spodoptera exigua. Gene. 2015; 557: 215–221. 10.1016/j.gene.2014.12.038 [DOI] [PubMed] [Google Scholar]
- 67.Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007; 131: 136–145. 10.1016/j.cell.2007.09.020 [DOI] [PubMed] [Google Scholar]
- 68.Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009; 6: 897–903. 10.1038/nmeth.1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi IK, Hyun S. Conserved microRNA miR-8 in fat body regulates innate immune homeostasis in Drosophila. Dev Comp Immunol. 2012; 37: 50–54. 10.1016/j.dci.2011.12.008 [DOI] [PubMed] [Google Scholar]
- 70.Jin H, Kim VN, Hyun S. Conserved microRNA miR-8 controls body size in response to steroid signaling in Drosophila. Genes Dev. 2012; 26: 1427–1432. 10.1101/gad.192872.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang X, Zheng Y, Cao X, Ren R, Yu X, Jiang H. Identification and profiling of Manduca sexta microRNAs and their possible roles in regulating specific transcripts in fat body, hemocytes, and midgut. Insect Biochem Mol Biol. 2015; 62: 11–22. 10.1016/j.ibmb.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang J, Ge X, Li Z, Wang Y, Song Q, Stanley DW, et al. MicroRNA-281 regulates the expression of ecdysone receptor (EcR) isoform B in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2013; 43: 692–700. 10.1016/j.ibmb.2013.05.002 [DOI] [PubMed] [Google Scholar]
- 73.Zhou Y, Liu Y, Yan H, Li Y, Zhang H, Xu J, et al. miR-281, an abundant midgut-specific miRNA of the vector mosquito Aedes albopictus enhances dengue virus replication. Parasit Vectors. 2014; 7: 488 10.1186/s13071-014-0488-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jagadeeswaran G, Zheng Y, Sumathipala N, Jiang H, Arrese EL, Soulages JL, et al. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics. 2010; 11: 52 10.1186/1471-2164-11-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Macedo LMF, Nunes FMF, Freitas CV, Pires CV, Tanaka ED, Martins JR, et al. MicroRNA signatures characterizing caste-independent ovarian activity in queen and worker honeybees (Apis mellifera L.). Insect Mol Biol. 2016; 25: 216–226. 10.1111/imb.12214 [DOI] [PubMed] [Google Scholar]
- 76.Kim K, Vinayagam A, Perrimon N. A rapid genome-wide microRNA screen identifies miR-14 as a modulator of Hedgehop signaling. Cell Rep. 2014; 7: 2066–2077. 10.1016/j.celrep.2014.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Varghese J, Cohen SM. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drasophila. Genes Dev. 2007; 21: 2277–2282. 10.1101/gad.439807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nelson C, Ambros V, Baehrecke EH. miR-14 regualtes autophagy during developmental cell death by targeting ip3-kinase 2. Mol Cell. 2014; 56: 376–388. 10.1016/j.molcel.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jayachandran B, Hussain M, Asgari S. Regulation of Helicoverpa armigera ecdysone receptor by miR-14 and its potential link to baculovirus infection. J Invertebr Pathol. 2013; 114: 151–157. 10.1016/j.jip.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 80.Chang Z, Tang N, Wang L, Zhang L, Akinyemi IA, Wu Q. Identification and characterization of microRNAs in the white-backed planthopper, Sogatella furcifera. Insect Sci. 2016; 23: 452–468. 10.1111/1744-7917.12343 [DOI] [PubMed] [Google Scholar]
- 81.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003; 301: 336–338. 10.1126/science.1085242 [DOI] [PubMed] [Google Scholar]
- 82.Rota-Stabelli O, Campbell L, Brinkmann H, Edgecombe GD, Longhorn SJ, Peterson KJ, et al. A congruent solution to arthropod phylogeny: phylogenomics, microRNAs and morphology support monophyletic Mandibulata. Proc Biol Sci. 2011; 278: 298–306. 10.1098/rspb.2010.0590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, et al. The deep evolution of metazoan microRNAs. Evol Dev. 2009; 11: 50–68. 10.1111/j.1525-142X.2008.00302.x [DOI] [PubMed] [Google Scholar]
- 84.Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, Klaus S, et al. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010; 463: 1084–1088. 10.1038/nature08744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ninova M, Ronshaugen M, Griffiths-Jones S. Conserved temperal patterns of microRNA expression in Drosophila support a developmental hourglass model. Genome Biol Evol. 2014; 6: 2459–2467. 10.1093/gbe/evu183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marco A, Hui J, Ronshaugen M, Griffiths-Jones S. Functional shifts in insect microRNA evolution. Genome Biol Evol. 2010; 2: 686–696. 10.1093/gbe/evq053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou H, Arcila M, Li Z, Lee E, Henzler C, Liu J, et al. Deep annotation of mouse iso-miR and iso-moR variation. Nucleic Acids Res. 2012; 40: 5864–5875. 10.1093/nar/gks247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Long QX, Hu JL, Huang AL. Deep Sequencing of the Hepatitis B Virus Genome: Analysis of Multiple Samples by Implementation of theIllumina Platform. Methods Mol Biol. 2017;1540: 211–218. 10.1007/978-1-4939-6700-1_17 [DOI] [PubMed] [Google Scholar]
- 89.Ragupathy R, Ravichandran S, Mahdi MS, Huang D, Domaratzki M, Cloutier S. Deep sequencing of wheat sRNA transcriptome reveals distinct temporal expression pattern of miRNAs in response to heat, light and UV. Sci Rep. 2016; 6: 39373 10.1038/srep39373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Andrews SV, Ladd-Acosta C, Feinberg AP, Hansen KD, Fallin MD. "Gap hunting" to characterize clustered probe signals in Illumina methylation array data. Epigenetics Chromatin. 2016; 9: 56 10.1186/s13072-016-0107-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang L, Yang HL, Tu ZC, Wang XL. High-Throughput Sequencing of Microbial Community Diversity and Dynamics during Douchi Fermentation. PLoS ONE. 2016; 11: e0168166 10.1371/journal.pone.0168166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meuti ME, Denlinger DL. Evolutionary links between circadian clocks and photoperiodic diapause in insects. Integr Comp Biol. 2013; 53: 131–143. 10.1093/icb/ict023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ikeno T, Tanaka SI, Numata H, Goto SG. Photoperiodic diapause under the control of circadian clock genes in an insect. BMC Biol. 2010; 8: 116 10.1186/1741-7007-8-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pegoraro M, Tauber E. The role of microRNAs (miRNA) in circadian rhythmicity. J Genet. 2008; 87: 505–511. [DOI] [PubMed] [Google Scholar]
- 95.Chen W, Liu Z, Li T, Zhang R, Xue Y, Zhong Y, et al. Regulation of Drosophila circadian rhythms by miRNA let-7 is mediated by a regulatory cycle. Nat Commun. 2014; 5: 5549 10.1038/ncomms6549 [DOI] [PubMed] [Google Scholar]
- 96.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014; 11: 809–815. 10.1038/nmeth.3014 [DOI] [PubMed] [Google Scholar]
- 97.Kagias K, Podolska A, Pocock R. Reliable reference miRNAs for quantitative gene expression analysis of stress responses in Caenorhabditis elegans. BMC Genomics. 2014; 15: 222 10.1186/1471-2164-15-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang Y, Zhang X, Chen Y, Guo J, Ling H, Gao S, et al. Selection of reference genes for normalization of microRNA expression by RT-qPCR in sugarcane buds under cold stress. Front Plant Sci. 2016; 7: 86 10.3389/fpls.2016.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000; 46: 69–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
34.7% was mapped to miRBase, 38.06% was mapped to the transcriptome of G. molesta and 27.24% were mapped to the genome of D. plexippus.
(TIF)
The average expression stability (M value) was calculated for each candidate and the sRNA with the lowest M value is considered as the most stably expressed reference gene.
(TIF)
Average pairwise variations (V values) were calculated between the normalization factors NFn and NFn+1, and the addition of reference gene is not required when the V value is below 0.15.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.