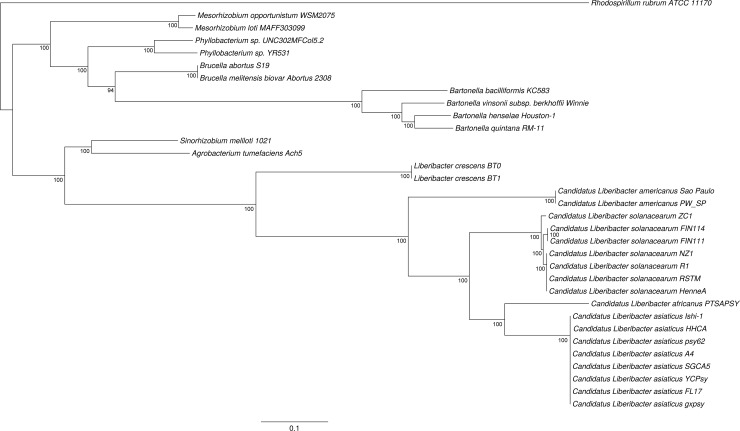
Fig 2. Phylogenetic tree constructed of 88 protein-coding genes from 33 bacterial genome datasets, belonging to genera ‘Candidatus Liberibacter’, Liberibacter, Agrobacterium, Sinorhizobium, Brucella, Bartonella, Mesorhizobium, Phyllobacterium and Rhodospirillum.
The 88 ortholog groups were aligned using Muscle v3.8.31, and these multiple alignments were trimmed and concatenated into a supermatrix. The best amino acid substitute model was determined using ProtTest v3.4.1. Maximum-likelihood tree was constructed using RAxML v8.2.0 with ‘PROTGAMMAIWAGF’ setting. The numbers shown next to the branches indicate the percentage of bootstrap support values (1000 replicates). The branch lengths indicate the evolutionary distance as the number of base substitutions per site.