Abstract
Overexpression of CDR1, an efflux pump, is one of the major mechanisms contributing to drug resistance in Candida albicans. CDR1 p-lacZ was constructed and transformed into a Saccharomyces cerevisiae strain so that the lacZ gene could be used as the reporter to monitor the activity of the CDR1 promoter. Overexpression of CaNDT80, the C. albicans homolog of S. cerevisiae NDT80, increases the β-galactosidase activity of the CDR1 p-lacZ construct in S. cerevisiae. Furthermore, mutations in CaNDT80 abolish the induction of CDR1 expression by antifungal agents in C. albicans. Consistently, the Candt80/Candt80 mutant is also more susceptible to antifungal drugs than the wild-type strain. Thus, the gene for CaNdt80 may be the first gene among the regulatory factors involved in drug resistance in C. albicans whose function has been identified.
The prevalence of fungal infections has increased significantly in the past few decades. Among the organisms causing these infections, Candida albicans is the most frequently isolated fungal pathogen in humans and has caused morbidity in seriously debilitated and immunocompromised hosts (6). Coincident with the increased use of antifungal drugs, the incidences of drug resistance have also increased (24, 33, 36). The limited variety of antifungal agents and emerging drug resistance highlight the need to identify potential targets and elucidate the molecular mechanisms involved in drug resistance for the development of new effective antifungal agents.
Overexpression of efflux pumps, either major facilitators or ATP binding cassette (ABC) transporters, has been shown to be one of the major mechanisms of drug resistance in clinical isolates (9, 18, 19). The CDR1 gene, which encodes an ABC efflux pump, is identified by complementation of the pdr5 mutant, which is hypersensitive to cycloheximide, chloramphenicol, and azole drugs, in Saccharomyces cerevisiae (25). Mutations in CDR1 in C. albicans resulted in increased susceptibilities to azole drugs (29), which is consistent with the observation that overexpression of CDR1 contributes to the drug resistance of clinical isolates of C. albicans (17, 36). The AP-1 site and the drug-responsive element of the CDR1 promoter have been reported to be the cis-regulatory elements (5, 26). Furthermore, the existence of trans-regulatory factors of CDR1 has also been suggested (26). However, the molecular mechanism and the gene network regulating the expression of CDR1 and drug resistance are poorly understood.
In this study, as in previous studies (15, 16), we have successfully used S. cerevisiae as a model to study C. albicans, despite the differences between these two organisms. We have identified one predicted transcription factor, CaNdt80, the C. albicans homolog of S. cerevisiae Ndt80, which is a meiosis-specific transcription factor in S. cerevisiae (2, 3) and which is involved in drug resistance through the regulation of CDR1 in C. albicans.
MATERIALS AND METHODS
Strains and media.
The S. cerevisiae strains, C. albicans strains, and plasmids used in this study are listed in Table 1, Table 2, and Table 3, respectively. Yeast peptone dextrose (YPD; 1% yeast extract, 2% peptone, and 2% dextrose) and synthetic dextrose (SD; 0.67% yeast nitrogen base without amino acids and with 2% dextrose) were prepared as described by Sherman (30).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| 10560-2B | MATahis3::hisG ura3-52 leu2::hisG | G. Fink laboratory collection |
| 10560-5B | MATα trp1::hisG ura3-52 leu2::hisG | G. Fink laboratory collection |
| SLO1 | MATahis3::hisG ura3-52 leu2::hisG ade3::CDR1p-lacZ-LEU2 | This study |
| SLO2 | MATahis3::hisG ura3-52, leu2::hisG ade3::CDR1p-lacZ-LEU2(pRS426) | This study |
| SLO3 | MATahis3::hisG ura3-52 leu2::hisG ade3::CDR1p-lacZ-LEU2(LOB44) | This study |
| SLO4 | MATahis3::hisG ura3-52 leu2::hisG ade3::CDR1p-lacZ-LEU2 | This study |
| SLO5 | MATahis3::hisG ura3-52 leu2::hisG ade3::CDR1p-lacZ-LEU2(LOB45) | This study |
TABLE 2.
C. albicans strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| SC5314 | Wild type | 8 |
| BWP17 | ura3Δ::λ imm434/ura3Δ::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 35 |
| DSY448 | ura3Δ::λ imm434/ura3Δ::λ imm434 cdr1::hisG/cdr1::hisG-URA3-hisG | 29 |
| YLO131 | ura3Δ::λ imm434/ura3Δ::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG CaNDT80/Candt80::GFP-Arg4 | This study |
| YLO132 | ura3Δ::λ imm434/ura3Δ::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG Candt80::GFP-Arg4/Candt80::URA3-dpl200 | This study |
| YLO133 | ura3Δ::λ imm434/ura3Δ::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG Candt80::GFP-Arg4/Candt80::URA3-dpl200 ENO1/eno1::ENO1-tetR-ScHAP4-3xHA-HIS1 | This study |
| YLO137 | ura3Δ::λ imm434/ura3Δ::λ imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG Candt80::GFP-Arg4/Candt80::URA3-dpl200:: CaNDT80::HIS1 | This study |
TABLE 3.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| B2159 | ADE3 URA3 CEN plasmid | G. Fink laboratory collection |
| pRS305 | LEU2 2μm plasmid | 31 |
| pRS315 | LEU2 CEN plasmid | 31 |
| pRS426 | URA3 2μm plasmid | 1 |
| YIp363 | β-Gal reporter LEU2-integrating plasmid | 22 |
| pGEM-HIS1 | CaHIS1-integrating plasmid | 35 |
| pRS-ARG4ΔSpeI | CaARG4 CEN plasmid | 35 |
| pDDB57 | CaURA3-dpl200 cassette | 34 |
| pGFP-HIS1 | GFP-CaHIS1 cassette | 7 |
| LOB42 | 1.2-kb CDR1 promoter-lacZ in- frame fusion in YIp363 | This study |
| LOB43 | 1.6-kb ADE3 in LOB42 | This study |
| LOB44 | CaNDT80 URA3 2μm plasmid | This study |
| LOB45 | 2.8-kb full length CaNDT80 URA3 2μm PRS426 plasmid | This study |
| LOB46 | NDT80 URA3 2μm plasmid | This study |
| LOB47 | GFP-CaARG4 cassette | This study |
| LOB48 | tetR-ScHAP4-3xHA CaHIS1- encoding plasmid | This study |
| LOB49 | CaNDT80 CaHIS1-integrating plasmid | This study |
Construction of CDR1p-lacZ strain.
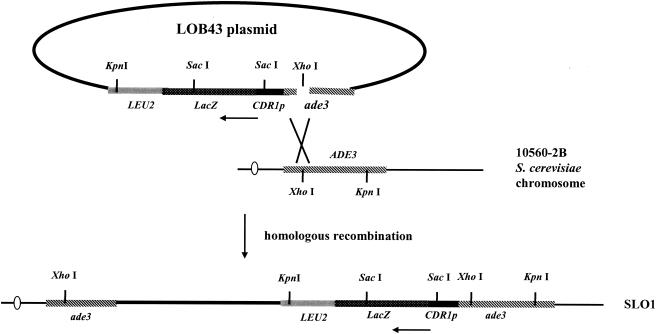
The CDR1p-lacZ fusion plasmid was constructed by using a DNA fragment generated by PCR from the promoter and the translation initiation codon ATG of CDR1 from C. albicans SC5314. The PCR fragment was generated with primers HJL21 (5′-dTTTCCCGGGGGATCCTCGTTACTCAA) and HJL22 (5′-dCCCAAGCTTGCATAATTTTTTTCTTTTTGACCT), which introduced 5′ XmaI and 3′ HindIII sites (underlined), respectively, for directional cloning into the XmaI and HindIII sites of plasmid YIp363. Plasmid LOB42 contained the CDR1p-lacZ in-frame fusion, in which lacZ is the reporter gene for monitoring the activity of the CDR1 promoter. Primers HJL44 (5′-TTTTCCCGGGCAGCAGTTTAGAAGCAAT) and HJL45 (5′-CCCCCCCGGGTGATTTGTCTTAACATT) were used to amplify the DNA fragment from the sequence 37 to 1648 bp downstream of the translation initiation site of the ADE3 gene. The fragment of the ADE3 gene was cloned in an antisense orientation into the XmaI site of the CDR1p-lacZ fusion of plasmid LOB42 to create plasmid LOB43. The LOB43 construct (Fig. 1) was digested with XhoI to linearize the DNA at position 337 bp downstream of the translation initiation site of the ADE3 gene and transformed into S. cerevisiae (10560-2B). The CDR1p-lacZ fusion of plasmid LOB43 was integrated into the ADE3 locus through homologous recombination to produce the Leu2+ transformant, strain SLO1.
FIG. 1.
Integration of the CDR1p-lacZ fusion in the ADE3 locus in S. cerevisiae. Horizontal arrows indicate the direction of transcription from the CDR1 promoter.
Screening for trans-regulators of CDR1 in S. cerevisiae.
A C. albicans genomic library constructed in high-copy-number S. cerevisiae plasmid 2μm-URA3 has been prepared (15) and was transformed into SLO1 containing the CDR1p-lacZ fusion. The lacZ gene was used as the reporter to monitor the activity of the CDR1 promoter. The β-galactosidase (β-Gal) activity was determined by both a filter assay and a liquid assay, as described previously (21). The library of transformants (approximately 500 colonies per 150-mm plate) was grown for 3 days before the transformants were replica plated onto filters laid on top of agar medium. When a Ura+ candidate strain containing a library plasmid had elevated β-Gal activity, as shown by a darker blue color than the control strain with only plasmid 2μm (1) in the filter assay, it was considered to harbor a candidate plasmid for the positive trans-regulatory factor of CDR1. When this was the case, the β-Gal activity of the strain would be reduced to the basal level when the plasmid was removed from the cells. Thus, the candidate strains were grown on YPD liquid medium for 2 days to lose the library plasmid. The β-Gal activities of the candidate plasmids and those of their counterparts which had lost the library plasmids were determined by β-Gal liquid assay.
Cloning of CaNDT80.
Primers HJL72 (5′-CGGGATCCTTGTGGCGATTTTCACTTTC) and HJL73 (5′-CCGGATCCTCAATGGGGGTGGATTGA) were used to amplify the genomic DNA containing the CaNDT80 gene of C. albicans from strain SC5314. The amplified DNA fragment starts from the position 578 bp upstream of the predicted start codon of the CaNDT80 gene to the position 479 bp downstream of the predicted stop codon (TAA) of CaNDT80. After digestion with BamHI, the DNA fragment was introduced into the pRS426 vector to generate plasmid LOB45.
Deletion of CaNDT80 in C. albicans.
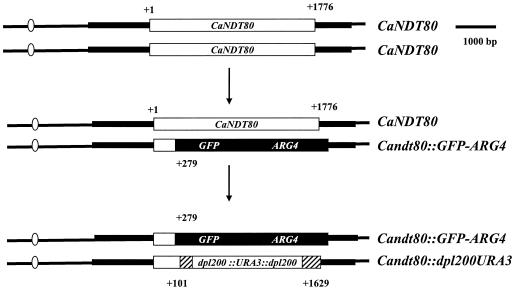
A homozygous Candt80/Candt80 mutant was constructed on the basis of the gene disruption method described previously (7, 34, 35), as shown in Fig. 2. The region from the position 279 bp downstream of the translation initiation site to the position 359 bp downstream of the stop codon of the CaNDT80 gene was replaced by GFP-ARG4. A DNA fragment containing the GFP-ARG4 construct flanked by short homologous regions (70 bp) of CaNDT80 at the two extremities was transformed into C. albicans strain BWP17. The region from the position 101 bp downstream of the translation initiation site to the position 148 bp upstream of the stop codon of the second copy of CaNDT80 was replaced by the URA3-dpl200-based cassette. A PCR product containing the URA3-dpl200 sequence with the CaNDT80 short homologous regions (70 bp) at the two extremities was transformed into YLO131 (CaNDT80/Candt80::GFP-ARG4). AccI-digested pT7tetR-HIS1 was integrated into the ENO1 promoter of YLO132, the Candt80/Candt80 homozygous mutant, to create strain YLO133. A BamHI DNA fragment containing wild-type CaNDT80 from plasmid LOB44 was inserted into the pGEM-HIS1 vector to generate pGEM-HIS1-CaNDT80, referred to as plasmid LOB49. Plasmid LOB49 was digested with SpeI, located in the promoter of CaNDT80, and was transformed into YLO132 to generate strain YLO137 (Candt80/Candt80::CaNDT80).
FIG. 2.
Construction of a null mutation in CaNDT80. The two copies of the CaNDT80 gene (open boxes) in the chromosome were sequentially replaced by the GFP-ARG4 construct (solid boxes) and the URA3-dpl200-based cassette (hatched boxes).
Antifungal susceptibility tests.
The Etest (23, 32) was used to determine the susceptibilities of S. cerevisiae strains containing either 2μm-CaNDT80 or plasmid 2μm to antifungal agents. Fluconazole (0.016 to 256 μg/ml) and ketoconazole (0.002 to 32 μg/ml) Etest strips (AB BIODISK, Solna, Sweden) were used. Homogenized colonies isolated from an overnight SD plate were transferred to 0.85% NaCl to achieve a density of 5 × 106 cells/ml. A sterile swab was dipped into the inoculum suspension and was then used to swab evenly the entire agar surface of an SD plate. The Etest strips were applied to the plate when the excess moisture had been absorbed completely. The agar dilution method was used to determine the susceptibilities of the C. albicans isolates to antifungal agents. Fluconazole and voriconazole were prepared to final concentrations of 25 and 1 μg/ml, respectively, in dimethyl sulfoxide (DMSO). Cells grown on medium containing an equal amount of DMSO in the absence of drug were used as controls. Strains were diluted to an optical density at 600 nm of 2 (approximately 2 × 107 cells/ml), and approximately 0.5 μl per spot was spotted onto plates containing different drugs with a replica plating device (Oxoid Inc., Nepean, Ontario, Canada). The strains were also serially diluted 10-fold.
Quantitative analysis of mRNA level by real-time PCR.
The C. albicans cells were harvested after they were grown in 20 ml of SD liquid medium in the absence or presence of 100 μg of miconazole per ml at 30°C for 1 h (optical density at 600 nm, 0.7 to 1.0). A real-time hot-start PCR was performed with an LC FastStartDNA Master SYBR Green I kit in a LightCycler instrument (catalog no. 2239264; Roche, Mannheim, Germany) to determine the level of mRNA. The real-time PCR was performed according to the instructions of the manufacturer. The expression of HGT4 in each strain was used as a control. The relative quantitation used two standard curves for the comparisons, and the results are given as a ratio (11). For the miconazole induction assay, the level of RNA isolated from different strains that had not been treated with miconazole was defined as 100%. The relative level of mRNA isolated from different strains that had been treated with miconazole was normalized to the level of mRNA isolated from the same strain that had not been treated with miconazole. To determine the effects of mutations in CaNDT80 after miconazole induction, the level of RNA isolated from the wild-type cells was defined as 100%.
RESULTS
Screening the trans-regulators of CDR1.
A total of approximately 24,000 independent library transformant colonies, which should cover about three times the C. albicans genome size, were screened. Of the 74 candidates picked initially, 16 were confirmed by the second filter assay to have β-Gal activity. Among these 16 candidates, 5 had higher β-Gal activities than the rest of the candidates and were chosen for further analysis. The higher β-Gal activities of two candidates resulted from mutations in a chromosome(s) in S. cerevisiae, since the strains still had the same level of β-Gal activity after they lost the plasmid. In contrast, the β-Gal activities of the remaining three candidates were reduced to the basal level when they lost the plasmid. Two of these three candidates harbored the same plasmid, named LOB44.
Overexpression of CaNDT80 increases the expression of CDR1p-lacZ.
S. cerevisiae strain SLO3 containing plasmid LOB44 had approximately sixfold higher β-Gal activity than strain SLO2 containing plasmid 2μm by the β-Gal liquid assay. Plasmid LOB44 contains a genomic insert of approximately 5 kb. This DNA fragment contains two full-length open reading frames. One is CaNdt80 and the other is orf6.1265, a short hypothetical protein of 106 amino acids (aa). The genomic insert was used for subcloning to produce LOB45, which harbored a 2.8-kb fragment with only the full-length gene of CaNDT80 generated by PCR. The β-Gal activity of parental strain SLO1 containing plasmid LOB45 (strain SLO5) was as high as that of the strain containing plasmid LOB44. These results demonstrate that the increased β-Gal activity of CDR1p-lacZ in S. cerevisiae is dependent on the presence of the CaNDT80 sequence.
CaNdt80 in C. albicans and Ndt80 in S. cerevisiae share the same DNA binding domain.
The CaNDT80 gene encodes a putative transcription factor. The transcription factor is 592 aa in length and shares the DNA binding domain with Ndt80, a meiosis-specific protein in S. cerevisiae. A comparison of the CaNdt80 and Ndt80 sequences is shown in Fig. 3. Ndt80 in S. cerevisiae is 672 aa. The sequence from amino acids 1 to 330 has been shown to be important for DNA binding activity. Two different domains have been identified. The residues from amino acids 1 to 58 are needed for sequence-specific interactions, and the residues from amino acids 59 to 330 contain a DNA binding domain. The sequence from amino acids 223 to 572 of Candt80 and those from amino acids 3 to the 330 of Ndt80 share 37.6% identity and 57.9% similarity (Fig. 3A). There was no similarity between the N terminus of CaNdt80 and the C terminus of Ndt80 (Fig. 3A).
FIG. 3.
Comparison of CaNdt80 in C. albicans and Ndt80 in S. cerevisiae. (A) Overall comparison of CaNdt80 and Ndt80. The regions shared between these two proteins are shown as thick gray bars. The domains different between those two proteins are presented as thin black lines. (B) Protein sequences of CaNdt80 and Ndt80. The CaNdt80 sequence is shown in boldface.
Overexpression of CaNDT80 decreases susceptibility to fluconazole and ketoconazole in S. cerevisiae.
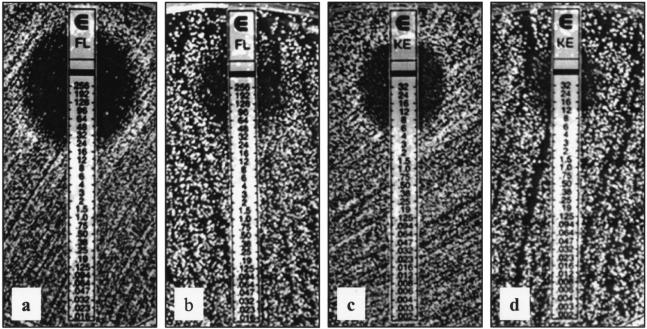
Overexpression of CaNDT80 increased the expression of CDR1p-lacZ, suggesting that CaNdt80 is an activator of CDR1 involved in drug resistance. According to the results of Etest, the overexpression of CaNDT80 decreased the susceptibility of S. cerevisiae to both fluconazole and ketoconazole (Fig. 4). The MIC of fluconazole increased from 24 to 64 μg/ml when cells overexpressed CaNDT80 (Fig. 4b). Consistently, the MIC of ketoconazole increased from 6 μg/ml to greater than or equal to 32 μg/ml when cells overexpressed CaNDT80 (Fig. 4d). The strain containing plasmid LOB45, which harbored CaNDT80, had similar susceptibilities to the two antifungal drugs. Thus, these data suggest that CaNdt80 of C. albicans is capable of activating the gene involved in drug resistance in S. cerevisiae.
FIG. 4.
Overexpression of CaNDT80 reduces S. cerevisiae susceptibilities to fluconazole (FL) and ketoconazole (KE). Antifungal susceptibility was determined by Etest. (a and b) Fluconazole strips; (c and d) ketoconazole strips; (a and c) S. cerevisiae with the 2μm-URA3 plasmid (1, 31) (SLO2); (b and d) S. cerevisiae with 2μm-URA3-CaNDT80 plasmid LOB45 (SLO3). The results were photographed after 3 days of growth at 30°C.
Mutations in CaNDT80 reduce the expression of CDR1 in C. albicans.
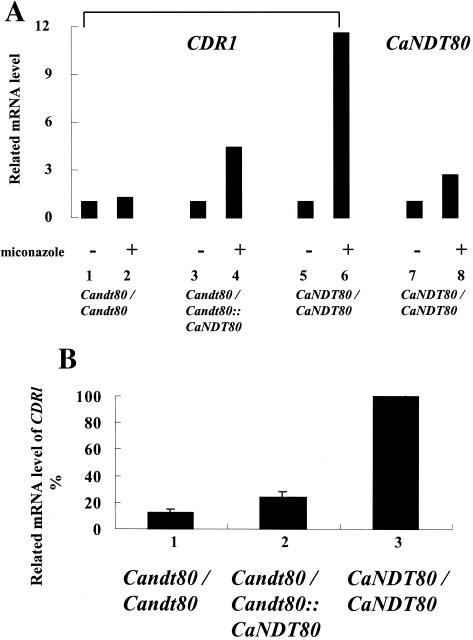
The CaNDT80 gene was expressed during vegetative growth in C. albicans (Fig. 5A, bar 7). The expression of CaNDT80 was increased by treating the cells with miconazole (Fig. 5A; compare bars 7 and 8). To elucidate the functions of CaNdt80 in C. albicans, we have constructed a Candt80/Candt80 homozygous mutant on the basis of the rapid gene disruption method (Fig. 2). No CaNDT80 mRNA was detected in the Candt80/Candt80 mutant, which therefore verified the presence of a null mutation in CaNDT80. As expected, CDR1 expression was also induced by miconazole treatment (Fig. 5A; compare bars 5 and 6). Homozygous null mutations in CaNDT80 abolished the induction of CDR1 expression in the presence of the drug (Fig. 5A; compare bars 1 and 2). This result is consistent with the idea that CaNdt80 is an activator of CDR1. However, CDR1 expression was not completely abolished by the null mutation in CaNDT80. In the presence of drugs, the level of CDR1 mRNA in the Candt80/Candt80 mutant strain was reduced to 15%, not 0%, of that in the wild-type strain (Fig. 5B; compare bars 1 and 3).
FIG. 5.
Mutations in CaNDT80 abolish the induction of CDR1 by miconazole. (A) Total RNA was isolated from cells that were not treated (bars 1, 3, 5, and 7) or cells that were treated with 100 μg of miconazole per ml (bars 2, 4, 6, and 8) for 1 h at 30°C. The level of RNA isolated from different strains without miconazole treatment was defined as 100%. The relative level of mRNA isolated from different strains with miconazole treatment was normalized to the level of mRNA isolated from the same strain without miconazole treatment. (B) Bar 1, Candt80/Candt80 (YLO133); bar 2, Candt80/Candt80::CANDT80 (YLO137); bar 3, CaNDT80/CaNDT80 (SC5314). The level of RNA isolated from wild-type cells was defined as 100%.
Mutations in CaNDT80 increase susceptibilities to antifungal agents in C. albicans.
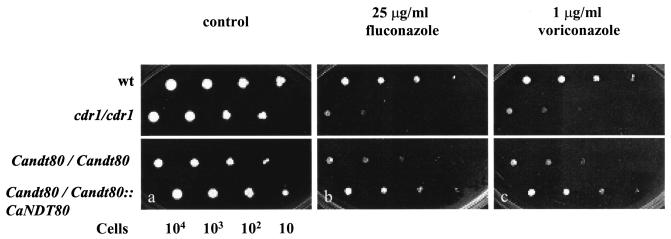
According to the results of the agar dilution assay, strains with mutations in either CDR1 or CaNDT80 were more susceptible to fluconazole and voriconazole (Fig. 6). Cells grew in all spots in the absence of drugs (Fig. 6). The Candt80/Candt80 mutant was more susceptible to drugs than the wild-type strain or the rescued strain containing a copy of wild-type CaNDT80. Few cdr1/cdr1 cells grew on medium with drugs inoculated with 103 cells, while few Candt80/Candt80 cells grew on medium with drugs inoculated with 102 cells (Fig. 6). Thus, the cdr1/cdr1 mutant was more susceptible to drugs than the Candt80/Candt80 mutant.
FIG. 6.
Mutations in CaNDT80 increase susceptibility to azole drugs in the agar dilution assay. Cells were grown on media in the absence of drug (control) and in the presence of fluconazole and voriconazole. The results for four strains, CaNDT80/CaNDT80 (SC5314), cdr1/cdr1 (DSY448), Candt80/Candt80 (YLO133), and Candt80/Candt80::CaNDT80 (YLO137) strains, were photographed after 2 days of growth at 30°C.
DISCUSSION
Potential targets of CaNdt80.
In the present study, we have found that CaNdt80 is involved in drug resistance through regulation of the expression of CDR1 in C. albicans. This observation is consistent with the findings from a previous report that the expression of CaNDT80 in C. albicans is increased approximately threefold by itraconazole treatment (4). Overexpression of a fusion protein containing the potential trans-activation domain of CaNdt80 (the sequence from amino acids 1 to 216) and the DNA binding domain of Ndt80 (the sequence from amino acids 1 to 330) (13, 20) induced the expression of CDR1p-lacZ in S. cerevisiae (data not shown). These data suggest that the DNA binding domains of Ndt80 and CaNdt80 may recognize the same DNA sequence since the DNA binding domain of Ndt80 can replace that of CaNdt80. Ndt80 is autoregulated and activates its targets through the midsporulation element (MSE) consensus site (gNCRCAAAA/T, where g indicates a probable G, N indicates any nucleo-tide, and R indicates A or G) (3). One perfect MSE (gNCRCAAAA/T) is located 572 bp upstream of the translation-initiating codon (ATG) of CaNDT80; and three potential MSEs (CRCAAA) are located 70, 122, and 461 bp upstream of the translation initiation site. It will be interesting to determine whether CaNdt80, like Ndt80 in S. cerevisiae, also regulates its own expression.
Null mutations in CaNDT80 abolish the induction of CDR1 expression by drugs. Consistently, the promoter of CDR1 contains three potential MSEs (CRCAAA) located 270, 438, and 835 bp upstream from the translation initiation site, respectively. The level of CDR1 mRNA in the rescued strain is lower than that in the wild-type strain. Furthermore, the rescued strain (Candt80/Candt80::CaNDT80) is also more susceptible to fluconazole than the wild-type strain. These data suggest a dosage effect (+/+ > +/− > −/−) of CaNDT80 in C. albicans, as has been documented in some other genes of C. albicans (12).
Overexpression of CaNDT80 in S. cerevisiae decreased the susceptibility of the organism to antifungal agents, suggesting that in S. cerevisiae CaNdt80 may activate the genes involved in drug susceptibility, including ERG11, PDR1, and TOP1, whose promoters have either perfect or potential MSEs
Transcription factors with the DNA binding domain of CaNdt80.
Ndt80 was identified as a meiosis-specific transcription factor required for S. cerevisiae to exit the pachytene stage (2, 3). Mutations in NDT80 in S. cerevisiae did not alter the susceptibility to antifungal agents (unpublished data), which is consistent with previous findings that Ndt80 is active in S. cerevisiae diploid cells only during meiosis. The Orf6.4742 gene was previously designated CaNDT80 because of the highly conserved region of the novel DNA binding domain between CaNdt80 and Ndt80.
In addition to CaNdt80 and Ndt80, several proteins from higher eukaryotes, including Neurospora crassa, Dictyostelium discoideum, Caenorhabditis elegans, Drosophila melanogaster, and humans, also contain sequences homologous to this novel DNA binding motif of CaNdt80 (20). Most metastatic tumors are more resistant to chemotherapeutic drugs than their primary counterparts, and many drug-resistant tumors are more invasive than nonresistant parental cells (14, 28). There seems to be a connection between the invasiveness (virulence) and the drug resistance of the tumor cells. Interestingly, C11orf9, a human transcription factor with a DNA binding domain similar to that of the novel DNA binding motif CaNdt80, is also highly expressed in invasive or metastatic tumor cells (10). Hence, this motif represents a new type of DNA binding domain and may consist of members with unique pathways.
The development of drug resistance and the limited variety of antifungal drugs available for clinical therapy are issues in treatments for infectious diseases. In the present study, we have shown that CaNdt80 regulates one of the drug resistance pathways of C. albicans. Hence, our findings may open a new doorway for the development and design of new effective agents for the treatment of microbial infections.
Acknowledgments
We thank G. Fink, C. Gale, A. Mitchell, H. Nakayama, and D. Sanglard for strains and plasmids. We also thank M. Y. Tsao and J. S. Wang for technical assistance. We also thank E. M. Ju for editing the manuscript.
REFERENCES
- 1.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 2.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 3.Chu, S., and I. Herskowitz. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685-696. [DOI] [PubMed] [Google Scholar]
- 4.De Backer, M. D., T. Ilyina, X. J. Ma, S. Vandoninck, W. H. Luyten, and H. Vanden Bossche. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 45:1660-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Micheli, M., J. Bille, C. Schueller, and D. Sanglard. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43:1197-1214. [DOI] [PubMed] [Google Scholar]
- 6.Edwards, E. J. J. 1990. Candida species, p. 1943-1958. In G. L. Mandell, R. G. Douglas, and J. E. Bennett (ed.), Principles and practice of infectious diseases. Churchill Livingstone, New York, N.Y.
- 7.Gerami-Nejad, M., J. Berman, and C. A. Gale. 2001. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18:859-864. [DOI] [PubMed] [Google Scholar]
- 8.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 9.Karababa, M., A. T. Coste, B. Rognon, J. Bille, and D. Sanglard. 2004. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob. Agents Chemother. 48:3064-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiemer, A. K., K. Takeuchi, and M. P. Quinlan. 2001. Identification of genes involved in epithelial-mesenchymal transition and tumor progression. Oncogene 20:6679-6688. [DOI] [PubMed] [Google Scholar]
- 11.Kofron, M., T. Demel, J. Xanthos, J. Lohr, B. Sun, H. Sive, S. Osada, C. Wright, C. Wylie, and J. Heasman. 1999. Mesoderm induction in Xenopus is a zygotic event regulated by maternal VegT via TGFbeta growth factors. Development 126:5759-5770. [DOI] [PubMed] [Google Scholar]
- 12.Kohler, J. R., and G. R. Fink. 1996. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc. Natl. Acad. Sci. USA 93:13223-13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamoureux, J. S., D. Stuart, R. Tsang, C. Wu, and J. N. Glover. 2002. Structure of the sporulation-specific transcription factor Ndt80 bound to DNA. EMBO J. 21:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang, Y., S. McDonnell, and M. Clynes. 2002. Examining the relationship between cancer invasion/metastasis and drug resistance. Curr. Cancer Drug Targets 2:257-277. [DOI] [PubMed] [Google Scholar]
- 15.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 16.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Ribot, J. L., R. K. McAtee, L. N. Lee, W. R. Kirkpatrick, T. C. White, D. Sanglard, and T. F. Patterson. 1998. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 42:2932-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marger, M. D., and M. H. Saier, Jr. 1993. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem. Sci. 18:13-20. [DOI] [PubMed] [Google Scholar]
- 19.Michaelis, S., and C. Berkower. 1995. Sequence comparison of yeast ATP-binding cassette proteins. Cold Spring Harbor Symp. Quant. Biol. 60:291-307. [DOI] [PubMed] [Google Scholar]
- 20.Montano, S. P., M. L. Cote, I. Fingerman, M. Pierce, A. K. Vershon, and M. M. Georgiadis. 2002. Crystal structure of the DNA-binding domain from Ndt80, a transcriptional activator required for meiosis in yeast. Proc. Natl. Acad. Sci. USA 99:14041-14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosch, H. U., R. L. Roberts, and G. R. Fink. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5352-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers, A. M., A. Tzagoloff, D. M. Kinney, and C. J. Lusty. 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45:299-310. [DOI] [PubMed] [Google Scholar]
- 23.Pfaller, M. A., D. J. Diekema, S. A. Messer, L. Boyken, and R. J. Hollis. 2003. Activities of fluconazole and voriconazole against 1,586 recent clinical isolates of Candida species determined by broth microdilution, disk diffusion, and Etest methods: report from the ARTEMIS Global Antifungal Susceptibility Program, 2001. J. Clin. Microbiol. 41:1440-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaller, M. A., R. N. Jones, G. V. Doern, H. S. Sader, S. A. Messer, A. Houston, S. Coffman, and R. J. Hollis. 2000. Bloodstream infections due to Candida species: SENTRY antimicrobial surveillance program in North America and Latin America, 1997-1998. Antimicrob. Agents Chemother. 44:747-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prasad, R., P. De Wergifosse, A. Goffeau, and E. Balzi. 1995. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27:320-329. [DOI] [PubMed] [Google Scholar]
- 26.Puri, N., S. Krishnamurthy, S. Habib, S. E. Hasnain, S. K. Goswami, and R. Prasad. 1999. CDR1, a multidrug resistance gene from Candida albicans, contains multiple regulatory domains in its promoter and the distal AP-1 element mediates its induction by miconazole. FEMS Microbiol. Lett. 180:213-219. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds, A., V. Lundblad, D. Dorris, and M. Keaveney. 1997. Yeast vectors and assays for expression of cloned genes, p. 13.6.1-13.6.6. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, N.Y. [DOI] [PubMed]
- 28.Sahin, A. A. 2000. Biologic and clinical significance of HER-2/neu (cerbB-2) in breast cancer. Adv. Anat. Pathol. 7:158-166. [DOI] [PubMed] [Google Scholar]
- 29.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1996. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 40:2300-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherman, F. 2002. Getting started with yeast. Methods Enzymol. 350:3-41. [DOI] [PubMed] [Google Scholar]
- 31.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tapia, C., E. Leon, and E. Palavecino. 2003. Antifungal susceptibility of yeasts by Etest. Comparison of 3 media. Rev. Med. Chil. 131:299-302. [PubMed] [Google Scholar]
- 33.Vanden Bossche, H., D. W. Warnock, B. Dupont, D. Kerridge, G. S. Sen, L. Improvisi, P. Marichal, F. C. Odds, F. Provost, and O. Ronin. 1994. Mechanisms and clinical impact of antifungal drug resistance. J. Med. Vet. Mycol. 32(Suppl. 1):189-202. [DOI] [PubMed] [Google Scholar]
- 34.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65-70. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, Y. L., and H.-J. Lo. 2001. Mechanisms of antifungal agent resistance. J. Microbiol. Immunol. Infect. 34:79-86. [PubMed] [Google Scholar]