Abstract
The ST2 gene was originally identified as a primary responsive gene induced by stimulation with growth factors and by oncogenic stress. The ST2 gene harbors two distinct promoters – a distal promoter and a proximal promoter. In this study, we identified a novel type of serum‐responsive element in the ST2 proximal promoter using reporter gene analysis; this element includes a possible responsive element for STAT family proteins. Indeed, enforced expression of constitutively active STAT3 activated this promoter element and induced the expression of ST2 gene products. Furthermore, an oncogenic Ras (G12V) mutant also caused the expression of ST2 gene products by utilizing the proximal promoter. We also clarified that activation of the ST2 promoter by either growth stimulation or oncogenic Ras was suppressed by the inhibitors for STAT3 and ERK pathways. Our observations strongly suggest the importance of STAT family and ERK pathways for the induction of ST2 gene products by cell growth stimulation.
Keywords: ERK pathway, promoter, Ras, ST2/IL1RL1, STAT3
Abbreviations
- ERK
extracellular signal‐regulated kinase
- NP‐40
Nonidet P‐40
- STAT
signal transducers and activator of transcription
ST2 (IL1RL1, also called T1) was originally identified as a primary responsive gene that was highly induced by growth stimulation and oncogenic Ras‐induced signaling in quiescent murine fibroblasts 1, 2. The ST2 gene produces two main mRNAs by alternative splicing 3; these mRNAs encode a soluble secreted protein (ST2) and a membrane bound protein (ST2L) 1, 4, 5. By structural analysis of the ST2 cDNAs, ST2 and ST2L exhibit similarity with the IL‐1 receptor (IL‐1R) suggesting that the ST2 gene products, especially membrane bound ST2L, induce similar signaling cascades to IL‐1R. After identification of IL‐33 as a specific ligand for ST2/ST2L, ST2L stimulated by IL‐33 was demonstrated to activate the transcription factor NF‐κB and the mitogen‐activated protein kinase (MAPK) family through common signaling molecules to IL‐1R 6, 7, 8, 9. On the other hand, secreted ST2 protein functions as a decoy receptor for IL‐33 and negatively regulates the signaling pathway of ST2L 10, 11. However, the functional relationship between growth stimulation and the expression of ST2 gene products is still unknown.
It was reported that the ST2 gene products are specifically expressed in type 2 helper T cells (Th2), but not in Th1, suggesting that ST2 gene products also play important roles in the immune response of Th2 12, 13, 14. Recent studies reported the expression of ST2 gene products and their physiological functions in immune/inflammation‐related cells 15, 16. Human and mouse ST2 genes have two alternative promoters followed by distinct noncoding first exons called E1a and E1b 17. In a human fibroblast cell line, ST2 gene expression was driven only by the proximal promoter. On the other hand, in the case of hematopoietic cells, both the distal and proximal promoters were utilized for the expression of ST2 and ST2L 17. These observations suggested that ST2 promoter usage depends on the cell type. In various hematopoietic cell lines, the GATA transcription factor family exhibits important roles in ST2 gene expression 18, 19. On the other hand, it has not been clarified yet how the proximal promoter is regulated, because various promoter elements have been identified in the proximal promoter of the ST2 gene 20, 21.
In the current study, we analyzed the regulatory mechanism of the expression of ST2 and identified a novel serum‐responsive element (SRE) in the proximal ST2 promoter. Furthermore, this promoter element includes a possible STAT family binding sequence, and the enforced expression of constitutively active mutant of STAT3 enabled activation of the proximal ST2 promoter.
Materials and methods
Cell culture and transfection
Human fibroblast TM12, murine fibroblast NIH‐3T3, and HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 2 mm glutamine, and 100 units each of penicillin and streptomycin. Transfection for the reporter gene assay was performed using the calcium phosphate method or Fugene 6 (Promega, Fitchburg, WI, USA). Murine thymoma cell line EL‐4 was cultured in RPMI1640 containing 10% FBS and 55 μm β‐mercaptoethanol. To induce differentiation into Th2‐like cells, EL‐4 cells were treated with 20 nm 12‐myristate 13‐acetate (PMA) and 1 mm dibutyryl cAMP (Bt2cAMP) for 24 h. UT‐7 cells were cultured in RPMI1640 containing 10% FBS and 1 ng·mL−1 GM‐CSF 22. Transfection was carried out by electroporation method.
Constructions of reporter gene vectors
DNA fragments containing the distal and proximal human ST2 promoters were obtained from genomic DNA extracted from UT‐7 cells by PCR using the primer sets described below. Primer sequences were (distal promoter) 5′‐TTTGGTACCAGAGGAAAACTAGGCTGTGC‐3′ (sense) and 5′‐TTTCTCGAGCCTCATTGGGTTGTACTTGAG‐3′ (antisense), (proximal promoter) 5′‐TTTGGTACCCATTGGGTGAGTGTGAGAATG‐3′ (sense) and 5′‐TTTCTCGAGCCAAACTGAAAGACAGAGGGGATG‐3′ (antisense). These fragments were subcloned into KpnI and XhoI sites of a promoter‐less luciferase vector (Promega). Other fragments containing various lengths of 5′‐flanking region of the ST2 gene were amplified by PCR and subcloned as described above.
Reporter gene analysis
Cells were transfected with luciferase vectors containing various lengths of the 5′‐flanking region of ST2, and pRL‐TK containing Renilla luciferase to standardize the efficiency of each transfection. After 48 h, cells were harvested and lysed in passive lysis buffer (Promega). The lysates were processed for luciferase assays. Luciferase activities were determined by measuring luminescence intensity using a Lumat LB 9507 (Berthold Japan, Tokyo, Japan). Luminescence intensities derived from the reaction of firefly luciferase were normalized with that of Renilla luciferase.
Electrophoretic mobility shift assay
Cells were stimulated with 10% FBS for the indicated periods. TM12 cells were harvested and lysed in 500 μL of buffer A (10 mm Hepes‐KOH [pH 7.5], 10 mm KCl, 0.1 mm EDTA, 0.1% NP‐40, 1 mm DTT, and 5 μg·mL−1 aprotinin) and nuclei were collected by centrifugation at 3000 g for 1 min at 4 °C. The nuclei were suspended in 100 μL of buffer C (50 mm Hepes‐KOH [pH 7.5], 420 mm KCl, 0.1 mm EDTA, 5 mm MgCl2, 2% [v/v] glycerol, 1 mm DTT, and 5 μg·mL−1 aprotinin) and mixed with gentle rotation for 30 min at 4 °C. Then, the samples were clarified by centrifugation 10 000 g for 15 min at 4 °C and the supernatant was collected and used as the nuclear extract. A quantity of 5 μg of nuclear extract was reacted with oligonucleotides derived from the proximal promoter of ST2, which was 32P‐labeled using T4 polynucleotide kinase (TOYOBO, Osaka, Japan) at 30 °C for 30 min. The double‐stranded oligonucleotide used as a probe derived from human ST2 promoter was as follows: 5′‐TGTCAACATCAAGAATTCTTAGTACATGAT‐3′ (region from −130 to −101 in the ST2 proximal promoter).
Prediction of the transcription factors activating the ST2 promoter
To clarify which transcription factors regulate ST2 promoter activity, the sequence of the fragment from −130 to −101 in the proximal promoter of the human ST2 gene was analyzed using the TFBIND website (http://tfbind.hgc.jp).
Retrovirus production and infection
Retroviruses were prepared as described previously 23. HEK293T cells were transfected with helper retrovirus plasmids together with pBabePuro and MSCV‐ires‐Puro encoding the indicated proteins. Viruses were harvested 24–60 h posttransfection, pooled, and stored on ice. Exponentially growing cells (1 × 105 cells per 60‐mm‐diameter culture dish) were infected twice at 2 h intervals with 2 mL of fresh virus‐containing supernatant in complete medium containing 1.0 μg·mL−1 polybrene (Sigma‐Aldrich, St. Louis, MO, USA). Infected cells were collected by puromycin selection.
Reverse transcription‐PCR
Total RNA was extracted using TRI reagent (Sigma‐Aldrich). Single‐stranded cDNA was synthesized by reverse transcription from 2 μg of total RNA using ReverTra Ace (TOYOBO). Quantitative PCR using a KAPA SYBR Fast qPCR kit (KAPA Biosystems, Wilmington, MA, USA) was performed in a LightCycler 96 (Roche Diagnostics, Indianapolis, IN, USA) with PCR cycles set at 94 °C for 10 s, 50 °C for 15 s, and 72 °C for 1 min. The nucleotide sequences of primers used for the quantitative PCR were as follows: ST2 (forward 5′‐CAAGAAGAGGAAGGTCGAAATG‐3′ and reverse 5′‐ATGTGTGAGGGACACTCCTTAC‐3′); and ST2L (forward 5′‐CAAGAAGAGGAAGGTCGAAATG‐3′ and reverse 5′‐AGCAACCTCAATCCAGAACAC‐3′). To analyze the promoter usage for ST2 gene expression, the expression of ST2 and ST2L was detected with forward primers complementary to the distal first exon (5′‐GAATAAAGATGGCTAGGACCTCTGG‐3′) or the proximal first exon (5′‐AATGAGACGAAGGAGCGCCAAGTAG‐3′), and the reverse primers were as described previously 19. PCR products were detected by staining agarose gels with ethidium bromide. For the analysis of promoter usage of the human ST2 gene, the same protocol was utilized with murine ST2, and the sequences of utilized primers were described previously 17.
Statistical analysis of data
In the case of reporter gene analysis, we performed the experiment individually three times, and showed the data. In the graph, error bar means standard deviation (SD, n = 3).
Results
Differential usage of the distal and proximal ST2 promoters in human fibroblastic and hematopoietic cell lines
As reported previously, human and mouse ST2 genes have two alternative promoters, the distal and proximal promoters, followed by distinct noncoding first exons, called E1a and E1b 17. To analyze the promoter usage for the expression of ST2 gene products, we constructed separate reporter gene plasmids harboring the distal and proximal human ST2 promoters (Fig. 1A). We transfected the reporter plasmids into human fibroblasts TM12 cells and hematopoietic UT‐7 cells, respectively. Then, from performed luciferase reporter gene assays, we observed that the proximal promoter region but not the distal promoter region was activated in serum‐stimulated human fibroblast TM12 cells (Fig. 1B). On the other hand, activation of both the distal and proximal promoters was observed in UT‐7 cells. Next, we verified the promoter usage for the expression of ST2 gene products by RT‐PCR. When the distal and proximal promoters were utilized for ST2 expression, the amplification of the respective PCR products including distal first exon (E1a) and proximal first exon (E1b) were detectable. As shown in Fig. 1C, the PCR product including only E1b but not E1a was amplified from the cDNA of TM12; therefore, the expression of ST2 was driven only by the proximal promoter in TM12 cells. On the other hand, both the distal and proximal promoters were utilized for the expression of ST2 in UT‐7 cells. These results were consistent with a previous report 17. It was also reported that the expression of ST2 was induced by an oncogenic Ras mutant in NIH‐3T3 cells 2. Next, we analyzed the promoter usage for the expression of ST2 gene products in transformed NIH‐3T3 cells. As shown in Fig. 1D, an oncogenic Ras mutant (Ras (G12V)) activated the proximal promoter but not the distal promoter. Furthermore, we investigated the usage of the ST2 promoter by performing RT‐PCR with the same procedure (Fig. 1C). Unlike the case of serum‐stimulated human TM12 cells, the oncogenic Ras mutant caused the expression of not only ST2 but also ST2L in NIH‐3T3 cells (Fig. 1E). However, in both cases for ST2 and ST2L, the PCR product including E1b only but not E1a was amplified, suggesting that the oncogenic Ras mutant utilizes the proximal promoter for the expression of ST2 gene products. We also performed an analysis with total RNA extracted from a mouse thymoma cell line, EL‐4, which differentiated in response to stimulation with PMA and Bt2cAMP. In differentiated EL‐4 cells, the transcripts of ST2 and ST2L contained E1a, but not E1b. These results suggested that the oncogenic Ras mutant utilizes the proximal promoter for the expression of ST2 gene products, and this seems to be mediated by a similar mechanism to ST2 expression in serum‐stimulated TM12 cells. These results concur with previous observations 17, and showed that these reporter plasmids are suitable for the analysis of ST2 promoters.
Figure 1.
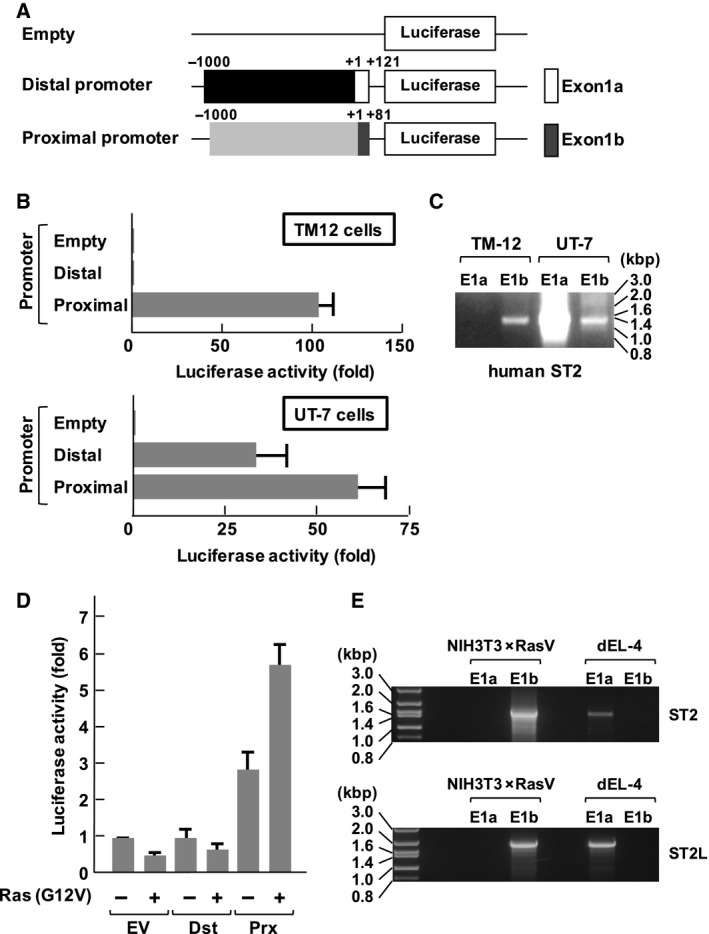
Differential usage of the distal and proximal ST2 promoters in human fibroblastic and hematopoietic cell lines. (A) The structures of luciferase vectors harboring the proximal promoters analyzed in this study are shown. An open box in the distal promoter shows the position of the distal promoter (E1a), and a gray‐colored box in proximal promoter shows the position of the proximal promoter (E1b). (B) Human fibroblasts TM12 cells and hematopoietic UT‐7 cells were transfected with the luciferase vectors shown in (A). Forty‐eight hours later, the cells were processed for reporter gene analysis. The promoter activities are shown in the graph. (C) Analysis of promoter usage of the human ST2 gene. Total RNA extracted from transformed TM‐12 and UT‐7 cells was analyzed by RT‐PCR using the primers described in the Materials and methods. The PCR products were separated on 1% agarose gels. (D) NIH‐3T3 cells were transfected with the combination of the indicated plasmids. Forty‐eight hours later, the cells were processed for reporter gene analysis. The promoter activities are shown in the graph. (E) Promoter usage of the murine ST2 gene was analyzed using total RNA extracted from transformed NIH‐3T3 × RasV and differentiated EL‐4 (dEL‐4) cells. The experiments were performed using the same method as (C). In the graph, error bar means standard deviation (SD, n = 3).
Identification of essential sequences in the ST2 proximal promoter that function in fibroblasts
We next tried to identify the functional promoter sequences for the expression of ST2 gene products by testing the promoter activity of a series of fragments of the proximal promoter (Fig. 2A). Trüb and colleagues reported that the enhancer element [12‐O‐tetradecanoylphorbol 13‐acetate (TPA)‐responsive element: TRE] was located 3.6 kbp upstream of the transcription initiation site in the proximal promoter of the murine ST2 gene 20. However, a reporter gene including 1 kbp upstream from the transcriptional initiation site, which lacked the reported TRE, still exhibited high promoter activity (Fig. 2B). As result, it was clarified that the promoter fragment from −330 to −101 in the proximal ST2 promoter was enough to exhibit promoter activity. In addition, deletion of E1b (Prox ‐1kbpΔE1b) significantly enhanced the promoter activity. Unexpectedly, the fragments of the ST2 proximal promoter lacking the reported enhancer elements were still activated, suggesting that critical enhancer elements for ST2 expression seem to be located in the region closer to the transcriptional initiation site.
Figure 2.
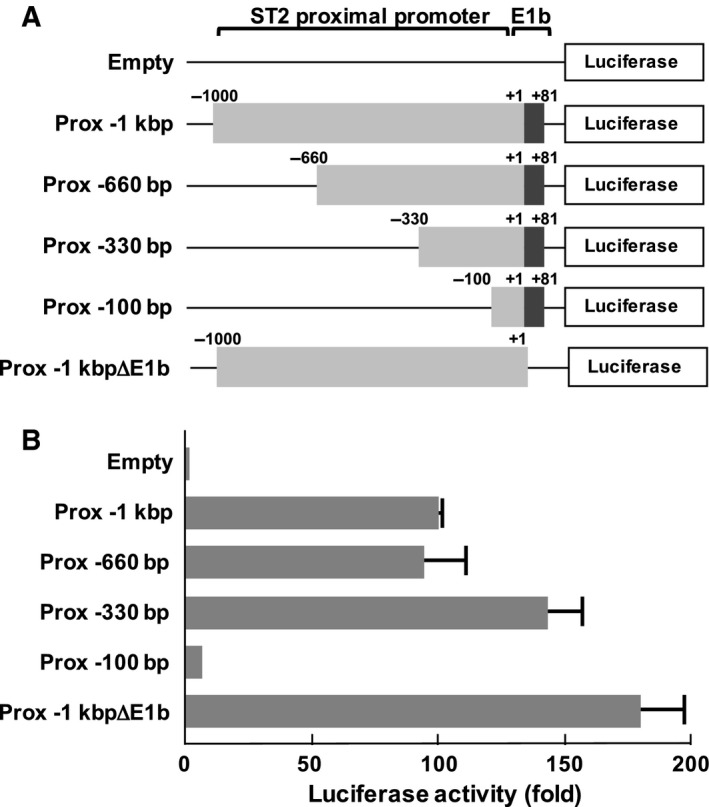
Upstream 12‐O‐tetradecanoylphorbol 13‐acetate‐responsive element and E‐box sequences are dispensable for activation of the proximal ST2 promoter. (A) The structures of luciferase vectors harboring proximal promoters analyzed in this study are shown. ‘Prox ‐1kbpΔE1b’ includes 1 kbp upstream from the transcriptional initiation site; however, it lacks Exon1b. The transcription initiation site is indicated as ‘+1’. The positions of promoter regions and E1b are indicated. (B) Human fibroblasts TM12 cells were transfected with the luciferase vectors shown in (A). Forty‐eight hours later, the cells were processed for reporter gene analysis. The promoter activities are shown in the graph. In the graph, error bar means standard deviation (SD, n = 3).
Finally, we found that the region from −130 to −101 was critical for promoter activity of ST2 in TM12 fibroblasts (Fig. 3A,B). We next performed an electrophoretic mobility shift assay (EMSA) using the promoter fragment from −130 to −101 in the proximal ST2 promoter and observed the presence of DNA–protein complexes, and the formation of these complexes was dependent on serum stimulation (Fig. 3C). Furthermore, this promoter element was required for full activation of the ST2 promoter in NIH‐3T3 cells transformed with Ras (G12V) (Fig. 3D). These findings clearly showed the importance of the region from −130 to −101 in the ST2 proximal promoter for the expression of ST2 gene products in human and murine fibroblasts.
Figure 3.
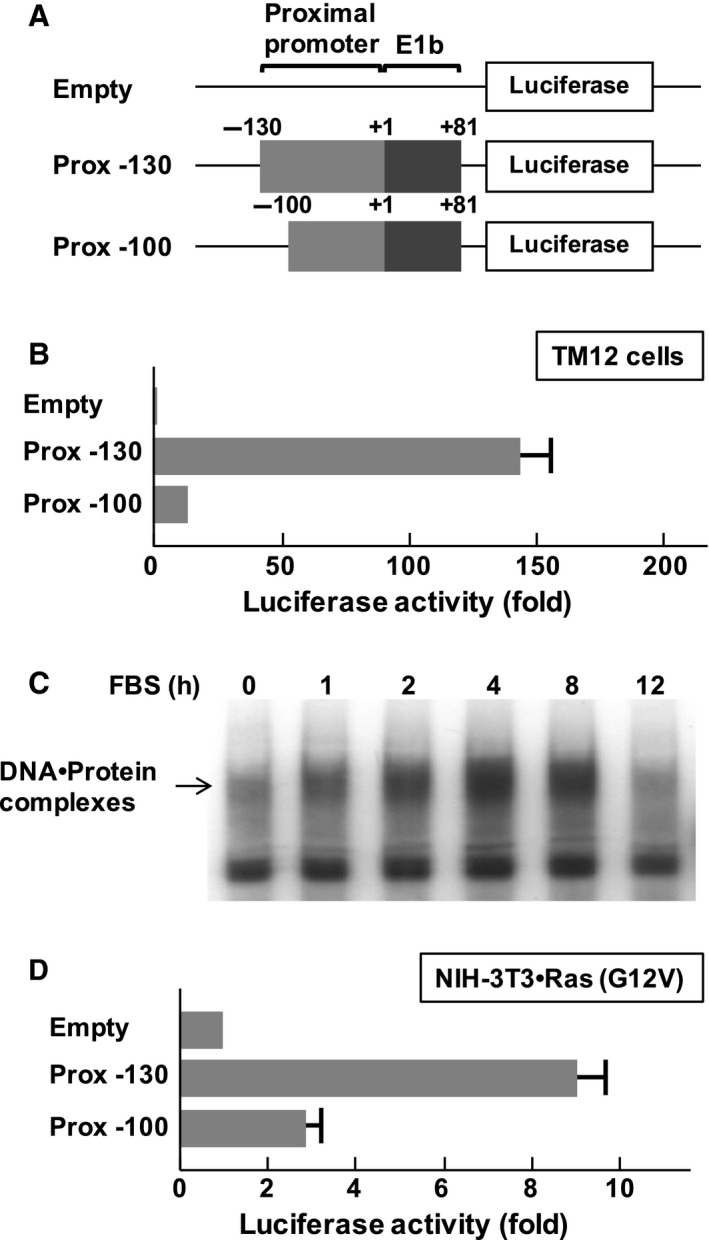
Identification of essential sequences in the ST2 proximal promoter in fibroblasts. (A) Structures of luciferase vectors analyzed in this study. Numbers indicate the position in the ST2 proximal promoter. The transcription initiation site is indicated as ‘+1’. The positions of promoter regions and E1b are indicated. (B) Human fibroblasts TM12 cells were transfected with the reporter gene plasmids. Forty‐eight hours later, the cells were harvested, and promoter activity was evaluated using luciferase assays. (C) After serum starvation, TM12 cells were stimulated with 10% fetal bovine serum for the indicated periods, and then nuclear extracts were prepared. Electrophoretic mobility shift assays (EMSAs) were performed with 32P‐labeled oligonucleotide probes containing the proximal promoter (−130 to −101). DNA–Protein complexes were separated on 5% nondenaturing polyacrylamide gels. (D) NIH‐3T3 cells were transfected with plasmids harboring Ras (G12V) and the indicated luciferase vectors. Forty‐eight hours later, the cells were processed for reporter gene analysis. In the graph, error bar means standard deviation (SD, n = 3).
Conserved regions in the human and mouse ST2 proximal promoters
When comparing the nucleotide sequences between the human and murine ST2 proximal promoters, it is possible to find two close regions of high homology starting approximately 150 bp upstream and ending 50 bp upstream of the transcription initiation site of the human ST2 gene. A direct alignment of this region is displayed in Fig. 4A. The direct alignment of the promoter sequences showed the presence of the conserved regions clarified by Harr plot analysis as sequences X and Y. In the previous study, conserved region X was shown to be a critical region for the activity of the ST2 proximal promoter in growing cells, although this promoter sequence failed to respond to serum stimulation 21. We next performed EMSA to test whether a cold nucleotide probe including the similar sequence from the mouse ST2 promoter could interrupt the formation of DNA–protein complexes with the promoter fragment from −130 to −101 in the human ST2 promoter. As shown in Fig. 4B, the DNA fragment of the mouse ST2 promoter effectively interrupted the formation of DNA–protein complexes as well as the fragment of the human ST2 promoter, suggesting that a critical transcription factor for ST2 expression seems to bind to the promoter fragment from −130 to −101 in the human/mouse ST2 promoters.
Figure 4.
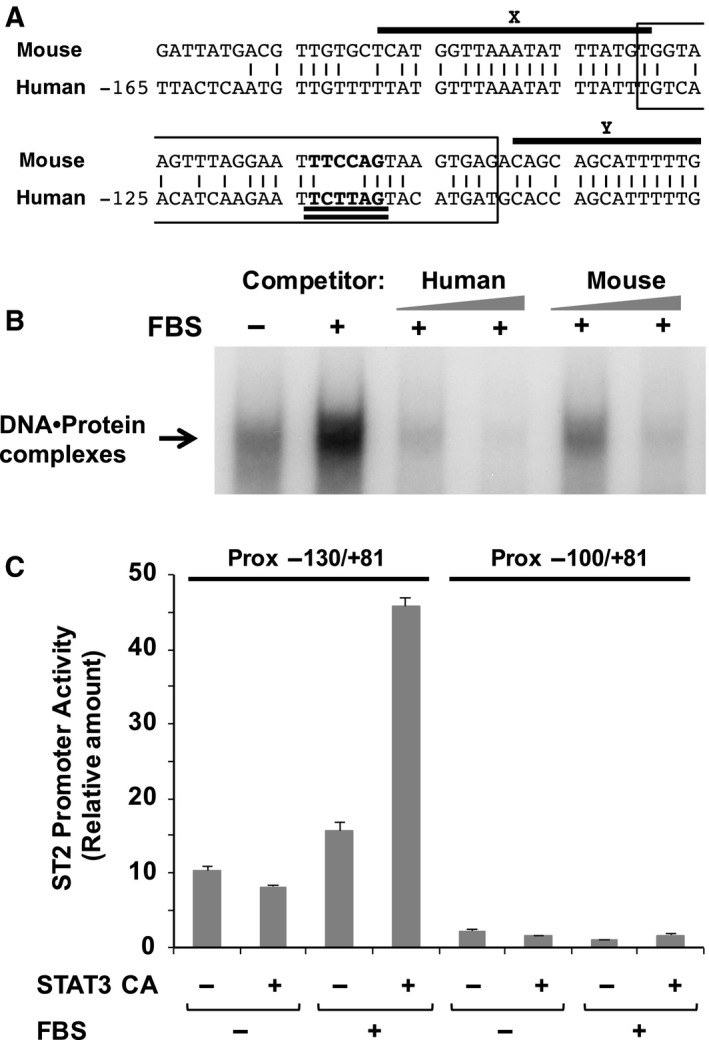
Conserved regions in the human and mouse ST2 proximal promoters. (A) The sequence similarity between the human and mouse ST2 proximal promoter is shown. The two upper‐lined sequences (X and Y) indicate the conserved regions clarified by Harr plot analysis. The boxed sequence indicates the nucleotide sequence of the probe utilized for EMSA. A possible consensus sequence for the STAT family is double underlined. (B) To show the effect of cold competitors derived from human and mouse ST2 proximal promoter on the formation of DNA–protein complexes, EMSA was performed as described in Fig. 3C and in the Materials and methods. (C) NIH‐3T3 cells were transfected with reporter gene plasmids with/without a constitutively active mutant of STAT3 (STAT3 CA). Forty‐eight hours later, the cells were harvested, and promoter activity was evaluated by performing luciferase assays. In the graph, error bar means standard deviation (SD, n = 3).
The presence of a STAT family responsive element in the ST2 proximal promoter
To clarify which transcription factors regulate ST2 promoter activity, we sought the transcription factor that is able to bind to the critical promoter sequences of both of the human and mouse ST2 genes by performing TFBIND analysis (see Materials and methods). We found a possible binding element for STAT family proteins in both the human and mouse ST2 promoter (Fig. 4A). Because STAT3 has been reported to lie downstream of the Ras signaling cascade 24, 25, we tested whether STAT3 activates the proximal promoter of the ST2 gene in murine fibroblast NIH‐3T3 cells. Strikingly, a constitutively active mutant of STAT3 stimulated the transcriptional activity from the −130/+81 fragment of the ST2 promoters but not from the −100/+81 fragment (Fig. 4C). STAT3‐mediated activation of the ST2 promoter required serum stimulation; however, STAT3 alone failed to enhance the promoter activity.
STAT3 and ERK pathways are involved in activation of the ST2 proximal promoter
To gain further insight, we next investigated the involvement of STAT3 in Ras (G12V)‐induced ST2 promoter activation. Napabucasin is a potent STAT3 inhibitor 26. With napabucasin treatment, the activity of the ST2 promoter was partially suppressed in NIH‐3T3 cells transformed with Ras (G12V) (Fig. 5A). Combining with the observation in Fig. 4C, it was suggested that STAT3 harbors the ability to activate the proximal ST2 promoter and is involved in Ras‐stimulated activation of the ST2 promoter. However, the ST2 promoter was still active when STAT3 was inactivated, suggesting that additional transcription factor(s) seem to be involved in activation of the ST2 promoter. It is well established that a Ras‐induced signal activates the ERK pathway through sequential activation of c‐Raf1 and MEK1/2 27. As shown in Fig. 5B, treatment with a MEK1/2 inhibitor, PD98059, caused drastic inhibition of ST2 promoter activity in TM12 cells. On the other hand, SB203580, an inhibitor of p38 MAPK exhibited no effect. Strikingly similar results were observed in the case of NIH‐3T3 cells transformed with Ras (G12V). These observations suggested the involvement of ERK in ST2 promoter activation induced by serum stimulation and an oncogenic Ras mutant.
Figure 5.
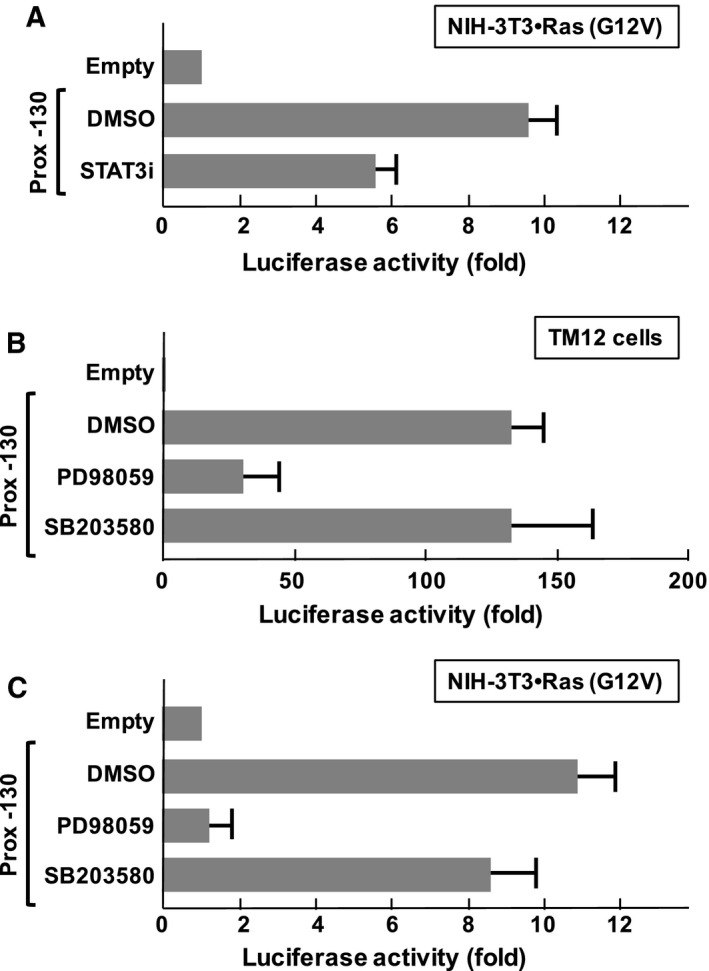
Functional involvement of STAT3 and ERK pathway in Ras (G12V)‐induced activation of ST2 promoter. (A) NIH‐3T3 cells were transfected with plasmids harboring Ras (G12V) and the indicated luciferase vectors. Then, to test the functional involvement of Ras‐related signaling molecules and STAT3, cells were treated with/without STAT3 inhibitor, 50 nm Napabucasin (STAT3i). Forty‐eight hours later, the cells were harvested, and promoter activity was evaluated by performing luciferase assays. (B) TM‐12 cells were transfected with reporter gene plasmids, and then 24 h later, cells were treated with indicated inhibitor compounds such as DMSO, 10 μm PD98059 or 10 μm SB203580. DMSO was utilized as a control. After 48 h, the cells were harvested, and promoter activity was evaluated by performing luciferase assays. (C) NIH‐3T3 cells were transfected with plasmids harboring Ras (G12V) and the indicated luciferase vectors. Then, cells were treated with the inhibitors indicated in (B). Forty‐eight hours later, the cells were harvested, and promoter activity was evaluated by performing luciferase assays. In the graph, error bar means standard deviation (SD, n = 3).
Discussion
A number of studies have suggested the functional involvement of ST2 gene products and their specific ligand IL‐33 in tumorigenesis and tumor metastasis of colorectal cancer and breast cancer 28, 29, 30, 31. On the other hand, it was also reported that the IL‐33/ST2L signaling axis exhibits an antitumor function 32, 33, suggesting that the functional direction of ST2 gene products seems to depend on the environmental conditions such as the tumor cell type. Additionally, we reported recently the functional importance of ST2 for cell cycle progression provoked by serum stimulation 34, and our study suggested that ST2 seems to be involved in cell proliferation of not only tumor cells but also normal fibroblasts. These observations emphasized the importance of investigation of the mechanism by which the ST2 promoter is regulated.
Previous studies reported the important role of a 148 bp enhancer element located 3.6 kbp upstream of the transcription initiation site in the proximal promoter of the murine ST2 gene, and suggested the presence of a TRE and three E‐box sequences in this region 20, 35. These elements could be activated by the cooperative actions of AP‐1 and basic helix‐loop‐helix transcription factors 36. However, this element was not sufficient for transcription of the ST2 gene. Additionally, in our study, we found that these elements seem to be dispensable for the activation of the ST2 proximal promoter by performing reporter gene analysis (Fig. 2).
In the current study, we identified a novel type of SRE in the proximal ST2 promoter, and this element includes a STAT family responsive element (Fig. 4A). The STAT family includes seven transcription factor proteins with high homology such as STAT1, STAT2, STAT3, STAT4, STAT5a and the closely related STAT5b, and STAT6 37. As shown in Fig. 4C, the enforced expression of STAT3 activated the proximal ST2 promoter; however, it still required serum stimulation, suggesting the requirement for additional factor(s). We found the nucleotides sequence 5′‐GGAATTTCC‐3′ in the essential region of the proximal ST2 promoter, and this is similar to the SRE. SRE is activated by the transcription factor SRF and Elk‐1 38, 39. As shown in Fig. 5B,C, ST2 promoter activation induced by serum stimulation and Ras (G12V) was effectively inhibited by treatment with PD98059, suggesting a requirement for the ERK pathway. It was reported that ERK phosphorylates and activates both STAT3 and Elk‐1 40, 41. Although we still have no direct evidence, STAT3 may cause full activation of the ST2 promoter mediated by the cooperation with Elk‐1 or SRF (Fig. 6). In addition, it is also possible that the ST2 proximal promoter could be activated by another family of STATs such as STAT5, and this possibility would need to be further investigated.
Figure 6.
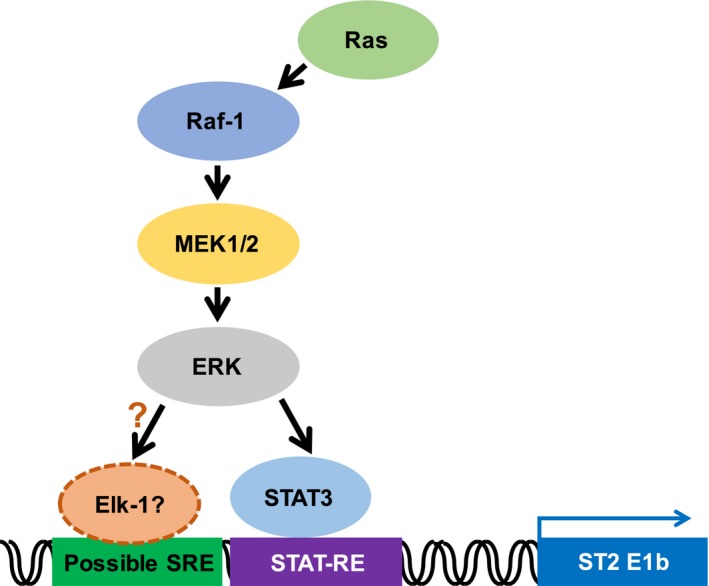
Working hypothesis how the Ras signal activates the ST2 promoter. Ras induces the expression of ST2 through the activation of the proximal promoter of ST2. The transcription factor STAT3 and ERK pathway are involved in the promoter activation.
Recently, we reported that an oncogenic Ras mutant induced the expression of not only ST2 but also IL‐33 in NIH‐3T3 cells 42. Although we have no evidence at present, the expression of ST2/ST2L and IL‐33 could be regulated synchronously. Combining these reports and our current findings together, it will be important to investigate the molecular mechanism of how the expression of ST2/ST2L and IL‐33 are regulated.
Conclusions
In the current study, we identified a novel type of SRE in the ST2 proximal promoter, which includes STAT family binding sequence. We also observed that STAT3 could activate this promoter sequence. This novel type of SRE in the ST2 proximal promoter also includes possible binding elements for SRF and Elk‐1. Using specific inhibitors, we clarified that the growth stimulation‐ and oncogenic Ras‐mediated ST2 promoter activation requires STAT3 and ERK pathway.
Author contributions
KT and ST designed the experiments in this study. KT and CA‐O performed the experiments. MF‐T and SO prepared reagents such as retroviral vectors and plasmids. KT, JM, and KY analyzed the data. KT, ST, and KY wrote the manuscript.
Acknowledgements
We thank Dr Scott Lowe for providing pBabePuro‐Ras (G12V). This work was supported by a Grant‐in‐Aid for Scientific Research from the MEXT (26440062 and 26460376), MEXT‐Supported Program for the Strategic Research Foundation at Private Universities (2013–2017), and JKA through its promotion funds from KEIRIN RACE.
References
- 1. Tominaga S (1989) A putative protein of a growth specific cDNA from BALB/c‐3T3 cells is highly similar to the extracellular portion of mouse interleukin 1 receptor. FEBS Lett 258, 301–304. [DOI] [PubMed] [Google Scholar]
- 2. Werenskiold AK, Hoffmann S and Klemenz R (1989) Induction of a mitogen‐responsive gene after expression of the Ha‐Ras oncogene in NIH 3T3 fibroblasts. Mol Cell Biol 9, 5207–5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Superti‐Furga G, Bergers G, Picard D and Busslinger M (1991) Hormone‐dependent transcriptional regulation and cellular transformation by Fos‐steroid receptor fusion proteins. Proc Natl Acad Sci USA 88, 5114–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yanagisawa K, Takagi T, Tsukamoto T, Tetsuka T and Tominaga S (1993) Presence of a novel primary response gene ST2L, encoding a product highly similar to the interleukin 1 receptor type 1. FEBS Lett 318, 83–87. [DOI] [PubMed] [Google Scholar]
- 5. Li H, Tago K, Io K, Kuroiwa K, Arai T, Iwahana H, Tominaga S and Yanagisawa K (2000) The cloning and nucleotide sequence of human ST2L cDNA. Genomics 67, 284–290. [DOI] [PubMed] [Google Scholar]
- 6. Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X et al (2005) IL‐33, an interleukin‐1‐like cytokine that signals via the IL‐1 receptor‐related protein ST2 and induces T helper type 2‐associated cytokines. Immunity 23, 479–490. [DOI] [PubMed] [Google Scholar]
- 7. Sanada S, Hakuno D, Higgins LJ, Schreiter ER, McKenzie AN and Lee RT (2007) IL‐33 and ST2 comprise a critical biomechanically induced and cardioprotective signaling system. J Clin Invest 117, 1538–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Funakoshi‐Tago M, Tago K, Hayakawa M, Tominaga S, Ohshio T, Sonoda Y and Kasahara T (2008) TRAF6 is a critical signal transducer in IL‐33 signaling pathway. Cell Signal 20, 1679–1686. [DOI] [PubMed] [Google Scholar]
- 9. Funakoshi‐Tago M, Tago K, Sato Y, Tominaga S and Kasahara T (2011) JAK2 is an important signal transducer in IL‐33‐induced NF‐κB activation. Cell Signal 23, 363–370. [DOI] [PubMed] [Google Scholar]
- 10. Hayakawa H, Hayakawa M, Kume A and Tominaga S (2007) Soluble ST2 blocks interleukin‐33 signaling in allergic airway inflammation. J Biol Chem 282, 26369–26380. [DOI] [PubMed] [Google Scholar]
- 11. Ohto‐Ozaki H, Kuroiwa K, Mato N, Matsuyama Y, Hayakawa M, Tamemoto H and Tominaga S (2010) Characterization of ST2 transgenic mice with resistance to IL‐33. Eur J Immunol 40, 2632–2642. [DOI] [PubMed] [Google Scholar]
- 12. Yanagisawa K, Kosaka A, Iwahana H, Nakanishi M and Tominaga S (1997) The expression of ST2 gene in helper T cells and the binding of ST2 protein to myeloma‐derived RPMI8226 cells. J Biochem 121, 95–103. [DOI] [PubMed] [Google Scholar]
- 13. Löhning M, Stroehmann A, Coyle AJ, Grogan JL, Lin S, Gutierrez‐Ramos JC, Levinson D, Radbruch A and Kamradt T (1997) T1/ST2 is preferentially expressed on murine Th2 cells, independent of interleukin 4, interleukin 5, and interleukin 10, and important for Th2 effector function. Proc Natl Acad Sci USA 95, 6930–6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coyle AJ, Lloyd C, Tian J, Nguyen T, Erikkson C, Wang L, Ottoson P, Persson P, Delaney T, Lehar S et al (1999) Crucial role of the interleukin 1 receptor family member T1/ST2 in T helper cell type 2‐mediated lung mucosal immune responses. J Exp Med 190, 895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Milovanovic M, Volarevic V, Radosavljevic G, Jovanovic I, Pejnovic N, Arsenijevic N and Lukic ML (2012) IL‐33/ST2 axis in inflammation and immunopathology. Immunol Res 52, 89–99. [DOI] [PubMed] [Google Scholar]
- 16. Sattler S, Smits HH, Xu D and Huang FP (2013) The evolutionary role of the IL‐33/ST2 system in host immune defence. Arch Immunol Ther Exp 61, 107–117. [DOI] [PubMed] [Google Scholar]
- 17. Iwahana H, Yanagisawa K, Ito‐Kosaka A, Kuroiwa K, Tago K, Komatsu N, Katashima R, Itakura M and Tominaga S (1999) Different promoter usage and multiple transcription initiation sites of the interleukin‐1 receptor‐related human ST2 gene in UT‐7 and TM12 cells. Eur J Biochem 264, 397–406. [DOI] [PubMed] [Google Scholar]
- 18. Nawijn MC, Dingjan GM, Ferreira R, Lambrecht BN, Karis A, Grosveld F, Savelkoul H and Hendriks RW (2001) Enforced expression of GATA‐3 in transgenic mice inhibits Th1 differentiation and induces the formation of a T1/ST2‐expressing Th2‐committed T cell compartment in vivo. J Immunol 167, 724–732. [DOI] [PubMed] [Google Scholar]
- 19. Hayakawa M, Yanagisawa K, Aoki S, Hayakawa H, Takezako N and Tominaga S (2005) T‐helper type 2 cell‐specific expression of the ST2 gene is regulated by transcription factor GATA‐3. Biochim Biophys Acta 1728, 53–64. [DOI] [PubMed] [Google Scholar]
- 20. Trüb T, Kalousek MB, Fröhli E and Klemenz R (1994) Growth factor‐mediated induction of the delayed early gene T1 depends on a 12‐O‐tetradecanoylphorbol 13‐acetate‐responsive element located 3.6 kb upstream of the transcription initiation site. Proc Natl Acad Sci USA 91, 3896–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tominaga S, Kato‐Yamazaki M, Yanagisawa K, Kawakami K and Tetsuka T (1994) The existence of a growth‐specific DNA binding factor for the promoter region of mouse ST2 gene. FEBS Lett 354, 311–314. [DOI] [PubMed] [Google Scholar]
- 22. Miura Y, Komatsu N and Suda T (1990) Growth and differentiation of two human megakaryoblastic cell lines; CMK and UT‐7. Prog Clin Biol Res 356, 259–270. [PubMed] [Google Scholar]
- 23. Tago K, Funakoshi‐Tago M, Itoh H, Furukawa Y, Kikuchi J, Kato T, Suzuki K and Yanagisawa K (2015) Arf tumor suppressor disrupts the oncogenic positive feedback loop including c‐Myc and DDX5. Oncogene 34, 314–322. [DOI] [PubMed] [Google Scholar]
- 24. Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD and Gutkind JS (2003) Epidermal growth factor receptor‐independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer Res 63, 2948–2956. [PubMed] [Google Scholar]
- 25. Park E, Park J, Han SW, Im SA, Kim TY, Oh DY and Bang YJ (2012) NVP‐BKM120, a novel PI3K inhibitor, shows synergism with a STAT3 inhibitor in human gastric cancer cells harboring KRAS mutations. Int J Oncol 40, 1259–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Y, Rogoff HA, Keates S, Gao Y, Murikipudi S, Mikule K, Leggett D, Li W, Pardee AB and Li CJ (2015) Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proc Natl Acad Sci USA 112, 1839–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kolch W (2005) Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol 6, 827–837. [DOI] [PubMed] [Google Scholar]
- 28. Jovanovic IP, Pejnovic NN, Radosavljevic GD, Pantic JM, Milovanovic MZ, Arsenijevic NN and Lukic ML (2014) Interleukin‐33/ST2 axis promotes breast cancer growth and metastases by facilitating intratumoral accumulation of immunosuppressive and innate lymphoid cells. Int J Cancer 134, 1669–1682. [DOI] [PubMed] [Google Scholar]
- 29. Yu XX, Hu Z, Shen X, Dong LY, Zhou WZ and Hu WH (2015) IL‐33 promotes gastric cancer cell invasion and migration via ST2‐ERK1/2 pathway. Dig Dis Sci 60, 1265–1272. [DOI] [PubMed] [Google Scholar]
- 30. Mager LF, Riether C, Schürch CM, Banz Y, Wasmer MH, Stuber R, Theocharides AP, Li X, Xia Y and Saito H et al (2015) IL‐33 signaling contributes to the pathogenesis of myeloproliferative neoplasms. J Clin Invest 125, 2579–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim JY, Lim SC, Kim G, Yun HJ, Ahn SG and Choi HS (2015) Interleukin‐33/ST2 axis promotes epithelial cell transformation and breast tumorigenesis via upregulation of COT activity. Oncogene 34, 4928–4938. [DOI] [PubMed] [Google Scholar]
- 32. Haga Y, Yanagisawa K, Ohto‐Ozaki H, Tominaga S, Masuzawa T and Iwahana H (2003) The effect of ST2 gene product on anchorage‐independent growth of a glioblastoma cell line, T98G. Eur J Biochem 270, 163–170. [DOI] [PubMed] [Google Scholar]
- 33. O'Donnell C, Mahmoud A, Keane J, Murphy C, White D, Carey S, O'Riordain M, Bennett MW, Brint E and Houston A (2016) An antitumorigenic role for the IL‐33 receptor, ST2L, in colon cancer. Br J Cancer 114, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tominaga S, Ohta S and Tago K (2016) Soluble form of the ST2 gene product exhibits growth promoting activity in NIH‐3T3 cells. Biochem Biophys Rep 5, 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kalousek MB, Trüb T, Schuermann M and Klemenz R (1994) T1 is a c‐Fos‐ and FosB‐responsive gene which is induced by growth factors through multiple signal transduction pathways. J Biol Chem 269, 6866–6873. [PubMed] [Google Scholar]
- 36. Kessler R, Laursen NB, Trüb T, Kalousek MB and Klemenz R (1997) Cooperative interactions of AP‐1 and basic helix‐loop‐helix transcription factors regulate T1 gene expression. Biol Chem 378, 657–667. [DOI] [PubMed] [Google Scholar]
- 37. Wake MS and Watson CJ (2015) STAT3 the oncogene – still eluding therapy? FEBS J 282, 2600–2611. [DOI] [PubMed] [Google Scholar]
- 38. Norman C, Runswick M, Pollock R and Treisman R (1998) Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c‐fos serum response element. Cell 55, 989–1003. [DOI] [PubMed] [Google Scholar]
- 39. Hipskind RA, Rao VN, Mueller CG, Reddy ES and Nordheim A (1991) Ets‐related protein Elk‐1 is homologous to the c‐fos regulatory factor p62TCF. Nature 354, 531–534. [DOI] [PubMed] [Google Scholar]
- 40. Lo RK, Cheung H and Wong YH (2003) Constitutively active Gα16 stimulates STAT3 via a c‐Src/JAK‐ and ERK‐dependent mechanism. J Biol Chem 278, 52154–52165. [DOI] [PubMed] [Google Scholar]
- 41. Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb MH and Shaw PE (1995) ERK phosphorylation potentiates Elk‐1‐mediated ternary complex formation and transactivation. EMBO J 14, 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ohta S, Tago K, Funakoshi‐Tago M, Matsugi J and Yanagisawa K (2016) Intracellular NF‐HEV/IL‐33 harbors essential roles in Ras‐induced cellular transformation by contributing to cyclin D1 protein synthesis. Cell Signal 28, 1025–1036. [DOI] [PubMed] [Google Scholar]


