Significance
The mitotic checkpoint system has an important role in ensuring correct segregation of chromosomes in mitosis. As long as not all chromosomes are attached correctly to the mitotic spindle, a mitotic checkpoint complex (MCC) is assembled and prevents chromosome separation by inhibiting the action of the ubiquitin ligase anaphase-promoting complex/cyclosome (APC/C). When the mitotic checkpoint system is turned off, MCC is disassembled to allow chromosome separation, but the mechanisms of MCC disassembly are poorly understood. Here we show that an important pathway in MCC disassembly is mediated by chaperonin containing TCP1, a complex previously known to play a role only in protein folding. The present study provides insight into molecular events responsible for mitotic checkpoint inactivation.
Keywords: cell cycle, ubiquitin, protein degradation
Abstract
The mitotic checkpoint system prevents premature separation of sister chromatids in mitosis and thus ensures the fidelity of chromosome segregation. When this checkpoint is active, a mitotic checkpoint complex (MCC), composed of the checkpoint proteins Mad2, BubR1, Bub3, and Cdc20, is assembled. MCC inhibits the ubiquitin ligase anaphase promoting complex/cyclosome (APC/C), whose action is necessary for anaphase initiation. When the checkpoint signal is turned off, MCC is disassembled, a process required for exit from checkpoint-arrested state. Different moieties of MCC are disassembled by different ATP-requiring processes. Previous work showed that Mad2 is released from MCC by the joint action of the TRIP13 AAA-ATPase and the Mad2-binding protein p31comet. Now we have isolated from extracts of HeLa cells an ATP-dependent factor that releases Cdc20 from MCC and identified it as chaperonin containing TCP1 or TCP1–Ring complex (CCT/TRiC chaperonin), a complex known to function in protein folding. Bacterially expressed CCT5 chaperonin subunits, which form biologically active homooligomers [Sergeeva, et al. (2013) J Biol Chem 288(24):17734–17744], also promote the disassembly of MCC. CCT chaperonin further binds and disassembles subcomplexes of MCC that lack Mad2. Thus, the combined action of CCT chaperonin with that of TRIP13 ATPase promotes the complete disassembly of MCC, necessary for the inactivation of the mitotic checkpoint.
The mitotic (or spindle assembly) checkpoint system ensures accuracy of chromosome segregation in mitosis by delaying anaphase until correct bipolar attachment of sister chromatids to the mitotic spindle is achieved (reviewed in refs. 1–3). When this checkpoint system is turned on, a mitotic checkpoint complex (MCC) accumulates, which inhibits the anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase, whose activity is necessary for the degradation of securin and thus for anaphase initiation (4, 5). MCC is composed of the checkpoint proteins Mad2, BubR1, and Bub3 associated with the APC/C activator Cdc20. The active checkpoint initiates the assembly of MCC by the conversion of Mad2 from an open (O-Mad2) to a closed (C-Mad2) conformation. C-Mad2 binds tightly to Cdc20, the Mad2–Cdc20 subcomplex associates with BubR1–Bub3 to form MCC and MCC binds tightly to the APC/C and inhibits its activity (1–3). During active mitotic checkpoint, about one-third of the MCC is bound to APC/C and the rest is in a pool of free MCC (6).
When the mitotic checkpoint is satisfied, the signal that inhibits the APC/C has to be eliminated to allow timely initiation of accurate chromosome segregation. This process is achieved by the rapid dissociation of both APC/C-bound and free pools of MCC. It has been reported that the release and disassembly of APC/C-bound MCC is accompanied by the ubiquitylation of Cdc20 (7), but it is not clear whether Cdc20 is the sole target of ubiquitylation (8, 9) and how ubiquitylation leads to MCC release. We have been studying the mechanisms of the disassembly of free MCC. It does not involve ubiquitylation (6, 10), but requires the energy of ATP hydrolysis (10). It was found that the different moieties of MCC are released by different ATP-requiring processes. Mad2 is released by the joint action of the TRIP13 AAA-ATPase and Mad2-binding protein p31comet (11, 12). The latter acts as a targeting protein for the binding of the ATPase to MCC (13, 14). The energy of ATP hydrolysis is apparently used in this process to promote a conformational change of Mad2 back from C-Mad2 to O-Mad2, leading to loss of its affinity for Cdc20 in MCC and thus to Mad2 release. By the action of TRIP13 and p31comet, MCC is converted to a remnant, called BC-1, in which Cdc20 is still associated with BubR1 (15). Cdc20 is released from BubR1 in MCC by a different process involving the phosphorylation of Cdc20 by Cdk1 (16). We noticed, however, that a considerable part of the release of Cdc20 from MCC was due to a phosphorylation-independent, but ATP-dependent process (16). The present study was initiated to elucidate the mechanisms of the latter process. We find that Cdc20 is dissociated from BubR1 in MCC and in its subcomplexes by the action of chaperonin containing TCP1 or TCP1–Ring complex (CCT/TRiC), a complex previously known to have roles in protein folding (reviewed in refs. 17–19). Along with the previously described pathways for MCC disassembly, the action of CCT/TRiC provides a mechanism for the complete dissociation of MCC into its free protein components and thus for the inactivation of the mitotic checkpoint.
Results
Isolation of an ATP-Dependent Factor That Promotes the Release of Cdc20 from Mitotic Checkpoint Complexes.
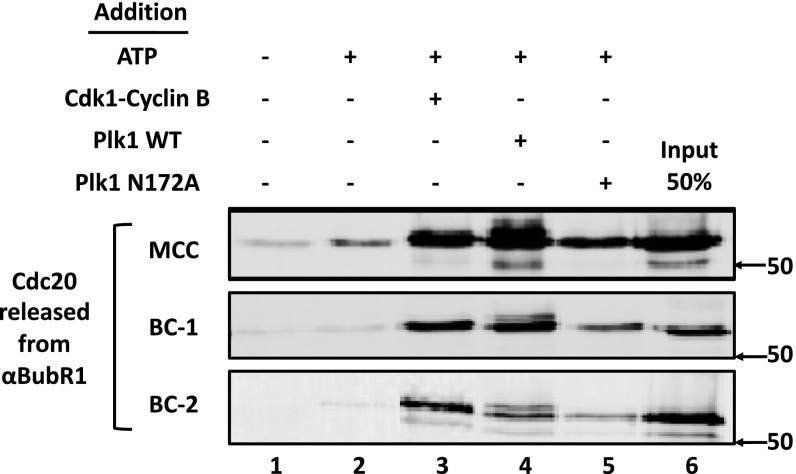
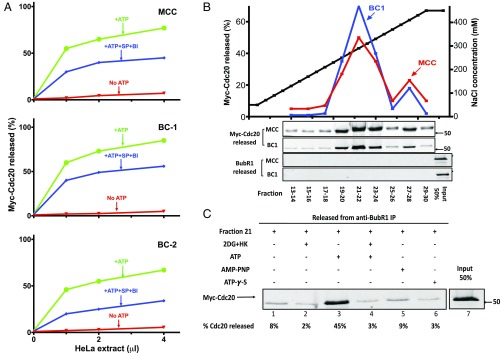
We have been studying the pathways of the disassembly of the mitotic checkpoint complexes (6, 10). The release of Mad2 is carried out by the AAA-ATPase TRIP13, jointly with the Mad2-binding protein p31comet (11), whereas a part of the dissociation of Cdc20 from BubR1 in MCC is promoted by Cdk-catalyzed phosphorylation of Cdc20 (16). We observed, however, that another part of Cdc20–BubR1 dissociation was not dependent on Cdk action (16). We now found that in a system reconstituted from purified components, protein kinase Plk1 (but not its catalytically inactive mutant N172A), also stimulates the release of Cdc20 from MCC (Fig. S1). Plk1 also acts on subcomplexes BC-1 (in which Cdc20 is bound to an N-terminal binding site of BubR1, ref. 15) and BC-2 (in which Cdc20 is bound to an internal site of BubR1, ref. 15) to dissociate Cdc20 from BubR1 (Fig. S1). To examine whether other processes, independent of phosphorylation by either Cdk or Plk, also participate in BubR1–Cdc20 dissociation, recombinant MCC and subcomplexes (all containing myc–Cdc20) bound to anti-BubR1 beads were incubated with extracts from nocodazole-arrested cells and additions as shown in Fig. 1A. Subsequently, the release of myc–Cdc20 to the supernatant was estimated. Essentially all release of Cdc20 from BubR1 depended upon the addition of ATP (Fig. 1A). Because extracts from checkpoint-arrested cells contain high levels of mitotic protein kinases, the ATP-dependent release of Cdc20 under these conditions represents the sum of phosphorylation-dependent and -independent processes. The addition of staurosporine (SP), which inhibits Cdks and most protein kinases (20), along with BI-2536 (BI), an inhibitor of Plks (21), inhibited only a part of the release of Cdc20 from MCC and its subcomplexes (Fig. 1A). These results suggested that a significant part of BubR1–Cdc20 dissociation may be carried out by an ATP-dependent, but phosphorylation-independent process.
Fig. S1.
Effects of Plk1 and Cdk on the disassembly of MCC and its subcomplexes. The release of Cdc20 from anti–BubR1-bound recombinant mitotic checkpoint complexes was determined as described in Materials and Methods. Where indicated, additions were at the following concentrations: ATP, 2 mM; Cdk1–cyclin B, 1,000 units; Plk1-WT and Plk1-N172A, 80 nM.
Fig. 1.
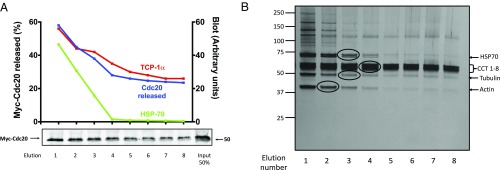
Isolation of an ATP-dependent disassembly factor from extracts of HeLa cells. (A) Part of the disassembly of MCC and of its subcomplexes by extracts of HeLa cells is ATP dependent, but phosphorylation independent. APC/C was removed from extracts of nocodazole-arrested HeLa cells by two sequential immunodepletions with anti-Cdc27, as described (10). Indicated volumes of this preparation were assayed for the disassembly of MCC and its subcomplexes, as described in Materials and Methods. Where indicated, the following additions were made: ATP, 2 mM; SP, 10 µM; and BI, 200 nM. Reactions without ATP were supplemented with hexokinase, 0.4 mg/mL and 2-deoxyglucose, 10 mM. (B) Anion exchange chromatography of ATP-dependent disassembly factor from extract of HeLa cells. Extracts from nocodazole-arrested HeLa cells (160 mg of protein) were applied to a 8-mL Source 15Q column (GE Healthcare) equilibrated with 50 mM Tris·HCl (pH 7.2), 50 mM NaCl, and 1 mM DTT. The column was washed with 30 mL of the above buffer and then subjected to a linear gradient of 50–450 mM NaCl (black squares) at a flow rate of 3.5 mL/min. Fractions of 8 mL were collected, concentrated by ultrafiltration, diluted 100-fold in a buffer consisting of 50 mM Tris·HCl (pH 7.2), 1 mM DTT, and 10% (vol/vol) glycerol, and concentrated again to a final volume of 1 mL. Samples of 1 μL of the indicated column fractions were assayed for the release of Cdc20 from MCC (red squares) and from BC-1 (blue squares) to the supernatant as described in Materials and Methods, in the presence of staurosporine and BI-2536. The Lower two panels are controls showing that BubR1 is not released from anti-BubR1 beads under the conditions of the disassembly activity assay, and therefore the release of Cdc20 is due to its dissociation from BubR1 in MCC. (C) The activity disassembly factor is dependent on ATP hydrolysis. Samples of 1 μL of anion exchange chromatography fraction 21 (B) were assayed for the release of Cdc20 from MCC bound to anti-BubR1 beads (Materials and Methods). Where indicated, additions were at the following concentrations: hexokinase (HK), 0.4 mg/mL; 2-deoxyglucose (2DG), 10 mM; ATP, AMP–PNP, and ATP-γ-S were supplemented at 2 mM.
To examine whether phosphorylation-independent disassembly activity is carried out by a protein that promotes ATP-requiring alterations in the complex, we searched for an ATP-dependent disassembly factor in extracts from HeLa cells. Extracts were subjected to chromatogaphy on an anion exchange Source Q column and disassembly activity was estimated in samples of column fractions by the release of myc–Cdc20 from MCC or BC-1, following incubation with ATP, staurosporine, and BI-2536. A major peak of activity eluted at about 300 mM NaCl of the salt gradient (Fig. 1B). The activity of the partially purified factor required ATP (Fig. 1C, lane 3). Disassembly activity was abolished when ATP was converted to ADP with hexokinase and deoxyglucose (Fig. 1C, lane 4) or when ATP was replaced by the analogs AMP–PNP [adenosine-5′-(β,γ-imido)-triphosphate] or ATP-γ-s [adenosine-5′-(γ-thio-)triphosphate] (Fig. 1C, lanes 5 and 6). These results suggested requirement of this factor for ATP hydrolyzable at the β−γ position.
Identification of the Disassembly Factor as CCT/TRiC Chaperonin.
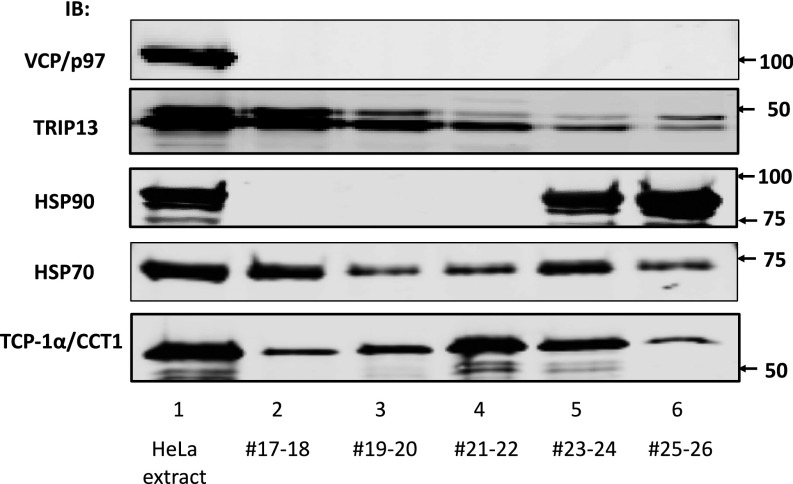
We tried to obtain some preliminary information about the identity of the disassembly factor by immunoblotting fractions of the anion exchange chromatography with antibodies available for AAA-ATPases, known to be involved in the disassembly of protein complexes (22) or chaperone proteins promoting changes in protein conformation. As shown in Fig. S2, the ATPases VCP/p97 or TRIP13 and the chaperone protein HSP90 did not elute in fractions 21 and 22 of the Source Q column, which contained the peak of the disassembly factor. The chaperone protein HSP70 eluted in two peaks, centered before (fractions 17 and 18) and after (fractions 23–34) of the peak of disassembly factor activity. By contrast, TCP1-α, a subunit of the CCT/TRiC chaperonin complex, eluted with a peak centered in fractions 21 and 22, similar to that of the disassembly factor (Fig. S2). These findings raised the possibility that the disassembly factor may be the CCT/TRiC chaperonin.
Fig. S2.
Coincidence of activity of releasing factor with CCT/TRiC chaperonin subunit, but not with some other chaperones. Samples of 0.5 μL of HeLa cell extract and 2 μL of the indicated anion exchange column fractions were immunoblotted with the indicated antibodies. Arrows on the Right indicate the positions of marker proteins (in kilodaltons).
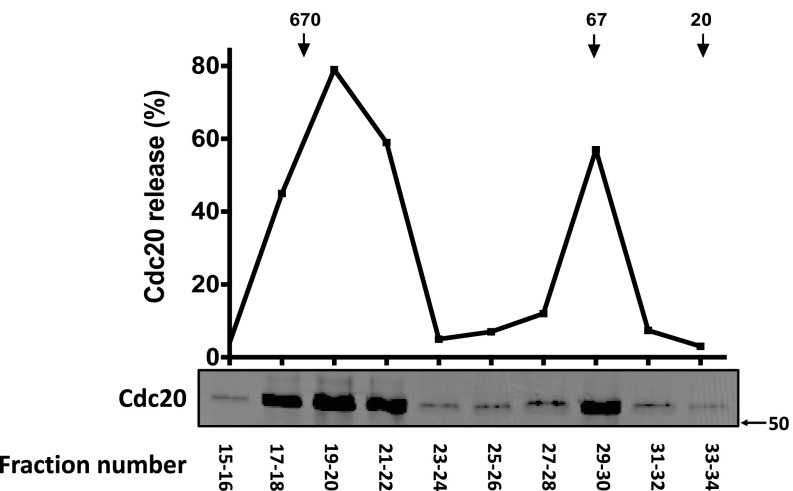
We tried to identify the ATP-dependent disassembly factor by its purification to near homogeneity. Further purification of the disassembly factor by gel filtration chromatography on Superdex 200 (Fig. S3) yielded a major peak of activity of a large complex eluting near the void volume and a minor peak at around 70 kDa. The major peak was subjected to affinity chromatography on ATP-6-AH agarose beads. The factor bound to the resin and was subsequently eluted with ATP. Elution with 10 mM ATP was not complete, even after eight consecutive elutions (Fig. 2A), indicating heterogeneity of binding affinities to the ATP beads. Samples of eluates of the ATP affinity chromatography were subjected to silver staining (Fig. 2B) and the indicated protein bands (circled) were identified by mass spectrometry. A group of major proteins of ∼60 kDa were identified as subunits 1–8 of the CCT/TRiC chaperonin complex. More minor proteins were identified as HSP70, tubulin and actin. Tubulin and actin are known to be associated with CCT/TRiC for their folding (17–19). HSP70 is unlikely to be the disassembly factor because most of it eluted earlier than disassembly activity in ATP affinity chromatography (Fig. 2A). By contrast, the pattern of elution of TCP1-α protein from the ATP resin was similar to that of disassembly activity (Fig. 2A). These findings indicated that the ATP-dependent factor that dissociates Cdc20 from mitotic checkpoint complexes is either the CCT/TRiC chaperonin, or a protein of very low abundance, which is tightly associated with the CCT/TRiC chaperonin.
Fig. S3.
Further purification of disassembly factor by gel filtration chromatography. Partially purified disassembly factor from pooled fractions 21 and 22 of anion exchange chromatography (2 mg of protein) were applied to a Superdex 200 Increase 10/300 GL column (GE Healthcare), equilibrated with 50 mM Tris⋅HCl (pH 7.2), 150 mM NaCl, 1 mM DTT, 10% (vol/vol) glycerol, and 0.1 mg/mL BSA. Fractions of 500 μL were collected at a flow rate of 0.5 mL/min. Samples of 3 µL of the indicated fractions were assayed for Cdc20 release from anti–BubR1-bound BC-1 as described in Materials and Methods. Arrows at the Top indicate the elution position of marker proteins (in kilodaltons).
Fig. 2.
Identification of disassembly factor as the CCT/TRiC chaperonin. (A) Final purification of disassembly factor from HeLa cells by ATP affinity chromatography. Partially purified disassembly factor from pooled fractions 17–21 of the gel filtration column (Fig. S3) was applied to 0.5 mL of 6-AH-ATP affinity resin (Jena Biosciences). ATP affinity chromatography was performed according to instructions of the manufacturer, except that additional 10 mM MgCl2 was added to the wash buffer, and 10 mM ATP with 10 mM MgCl2 was added to the elution buffer. Eight consecutive elutions of 1 mL were collected and concentrated fourfold by ultrafiltration. Disassembly of MCC (Materials and Methods) was assayed in samples of 0.5-μL eluates and samples of 1 μL were blotted for TCP-1α and HSP70. (B) Silver staining of ATP affinity chromatography eluates of disassembly factor. Samples of 5 μL of the indicated ATP-affinity eluates (Fig. 2A) were electrophoresed on 4–12% Bis-Tris protein gels (Nupage, Invitrogen). The marked protein bands (encircled) were excised and subjected to tryptic digestion and mass spectrometric analysis. Identifications of proteins are shown on the Right, and electrophoretic migration positions of marker proteins (in kilodaltons) are shown on the Left.
Recombinant CCT5 Chaperonin Complex Has MCC-Dissociating Activity.
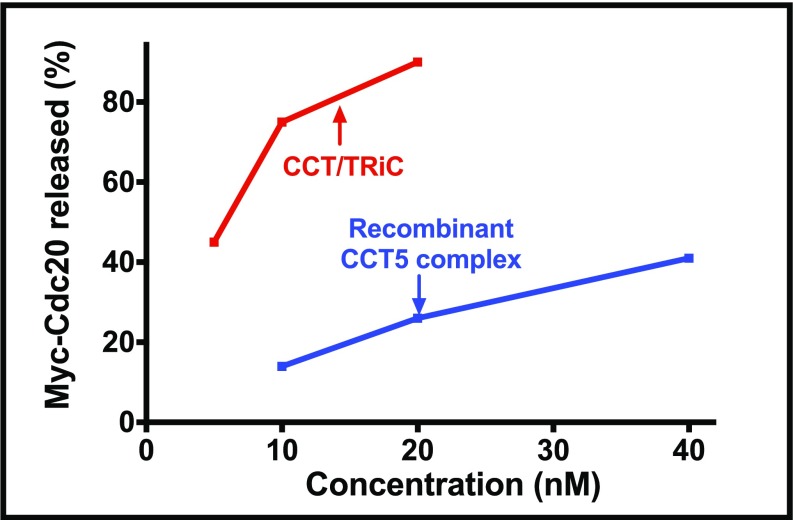
Trying to validate whether or not CCT/TRiC chaperonin is the MCC disassembly factor, we took advantage of the finding by King and coworkers (23) that homooligomers of recombinant CCT4 or CCT5 chaperonin subunits have biological activity in the refolding of proteins. We expressed human CCT5 in bacteria and separated its high molecular weight homooligomer by gel filtration on Superose 6. As shown in Fig. 3A, the CCT5 oligomer promoted the release of Cdc20 and of Mad2 from MCC. This suggested that the CCT chaperonin itself, and not a low-abundance protein from HeLa cells associated with the chaperonin, is the disassembly factor.
Fig. 3.
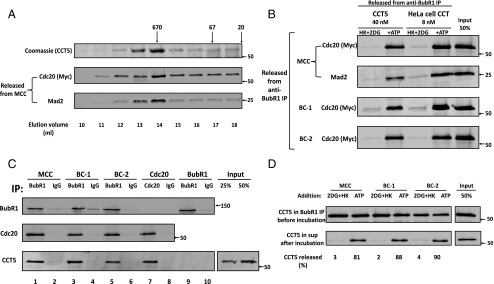
Activity and properties of recombinant CCT5 chaperonin complex in MCC disassembly. (A) Separation of recombinant CCT5 homooligomer by gel filtration chromatography. The pET21b plasmid-containing human his6–CCT5 gene insert (generously provided by Jonathan A. King, Massachusetts Institute of Technology, Cambridge, MA) was transformed into BL21 (DE3), and was expressed and purified on Ni-NTA agarose, as described (23). This material was applied to a Superose 6 10/300 GL size exclusion column (GE Healthcare) equilibrated with 20 mm Hepes/KOH, pH 7.4, 300 mm NaCl, 10 mm MgCl2, 5% (vol/vol) glycerol, and 1 mM DTT. Fractions of 0.5 mL were collected at a flow rate of 0.5 mL/min. Samples of 10 μL of column fractions at the indicated elution volumes were subjected to SDS/PAGE and Coomassie staining. Samples of 3 µL of the indicated fractions were analyzed for MCC disassembly as described in Materials and Methods, and released material was immunoblotted for myc–Cdc20 and Mad2. Arrows at the Top indicate the elution position of molecular mass marker proteins (in kilodaltons). (B) Comparison of dissociation of MCC, BC-1, and BC-2 by recombinant CCT5 complex and by purified HeLa cell CCT/TRiC chaperonin. Peak fractions of recombinant homooligomeric CCT5 complex and of purified heterooligomeric HeLa cell CCT/TRiC shown in Figs. 3A and 2B, respectively, were pooled and concentrated. The disassembly of various recombinant mitotic complexes by indicated concentrations of these preparations was assayed as described in Materials and Methods. Where indicated, additions were as follows: ATP, 2 mM, together with 10 mM phosphocreatine and 100 μg/mL creatine phosphokinase and HK, 400 μg/mL, together with 10 mM 2DG. Arrows on the Right indicate the positions of marker proteins (in kilodaltons). (C) CCT5 complex binds MCC, its subcomplexes and free Cdc20, but not free BubR1. The indicated mitotic checkpoint complexes were added to the assay at 100 nM in the presence of 100 nM purified CCT5 complex. Reaction mixtures contained in a volume of 25 μL ingredients at concentrations similar to those described for the assay of dissociation of complexes (Materials and Methods), except that ATP was omitted and 2-deoxyglucose and hexokinase were added. Following rotation at 4 °C for 2 h, samples were mixed with 5 μL of the indicated antibody-bound beads and subjected to immunoprecipitation with similar rotation. Immunoprecipitated material was then washed and analyzed as described in Fig. S5. Arrows on the Right indicate the positions of marker proteins (in kilodaltons). (D) Release of CCT5 from mitotic checkpoint complexes is ATP dependent. The indicated mitotic checkpoint complex proteins were first bound to CCT5 complex in the absence of ATP, as described in Fig. 3C. Subsequently, samples were immunoprecipitated with anti-BubR1 beads, beads were washed and then incubated (Materials and Methods) as indicated with hexokinase and 2-deoxyglucose (2DG+HK) or ATP. The release of CCT5 from anti-BubR1 beads was estimated and expressed as the percentage of initial CCT5 bound. Arrows on the Right indicate the positions of marker proteins (in kilodaltons).
Comparison of the disassembly activity of recombinant CCT5 complex with that of native CCT/TRiC chaperonin purified from HeLa cells (Fig. 3B) showed that both had an absolute requirement for ATP and both acted not only on MCC, but also on its subcomplexes BC-1 and BC-2. This finding indicated again the similarity of the properties of recombinant CCT5 complex with those of CCT/TRiC. It should be noted, however, that the specific activity of the homooligomer in MCC dissociation was five- to eightfold lower than that of CCT/TRiC purified from HeLa cells (Fig. 3B and Fig. S4). This finding could be due to incomplete folding of bacterially expressed CCT5 or to better function of some specific subunits of HeLa cell chaperonin in its action in the disassembly of MCC.
Fig. S4.
Disassembly activities of recombinant CCT5 complex vs. HeLa cell CCT/TRiC. The activities of indicated concentrations of either recombinant CCT5 complex or of purified HeLa cell CCT/TRiC in the disassembly of MCC were assayed as described in Materials and Methods.
Mode of Action of CCT Chaperonin in MCC Disassembly.
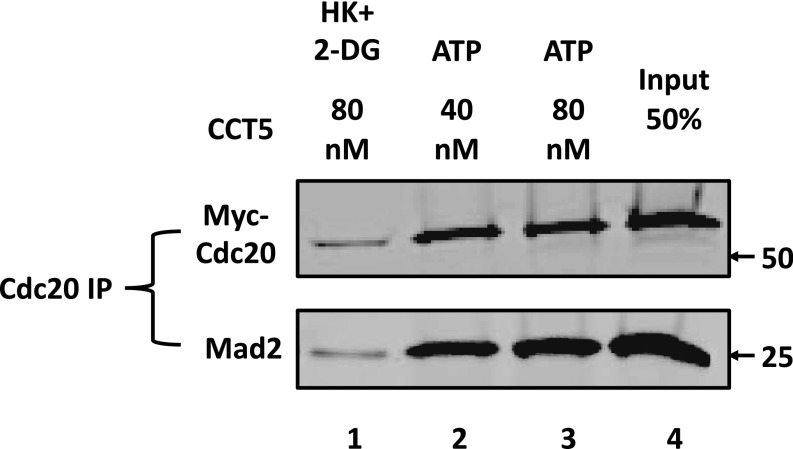
If CCT chaperonin acts mainly in the dissociation of Cdc20 from BubR1, it could be expected that released Cdc20 is still associated with C-Mad2, to which it is tightly bound (1–3). In the experiment shown in Fig. S5, we examined this question by coimmunoprecipitation of Mad2 with Cdc20 in material released from MCC. Indeed, anti-Cdc20 immunoprecipitate of released material contained not only Cdc20, but also Mad2, indicating the release of a Cdc20-Mad2 subcomplex in this process.
Fig. S5.
CCT complex releases Mad2–Cdc20 subcomplex from MCC. MCC disassembly by CCT5 complex at the indicated concentrations was carried out as described in Materials and Methods. Released material was passed through 0.45-μM centrifugal filters and subjected to immunoprecipitation with anti-Cdc20 beads by rotation at 4 °C for 2 h. The beads were then washed three times with a buffer consisting of 50 mM Tris⋅HCl (pH 7.2), 20% (vol/vol) glycerol, 300 mM NaCl, and 1% (vol/vol) Nonidet P-40. Immunoprecipitates were resolved by SDS/PAGE and immunoblotted for Myc–Cdc20 and Mad2. Where indicated, additions were as described in Fig. 3B. Arrows on the Right indicate the electrophoretic migration of marker proteins (in kilodaltons).
We also asked how are mitotic checkpoint complexes bound to CCT chaperonin. In the experiment shown in Fig. 3C, the binding of different checkpoint complexes to CCT in the absence of ATP was determined. CCT5 homooligomer bound strongly not only to MCC but also its subcomplexes BC-1 and BC-2 (lanes 1, 3, and 5). All these complexes contain Cdc20 and BubR1, so it was possible that the chaperonin binds to the Cdc20 moiety, to the BubR1 moiety, or to both. In agreement with a previous report (24), we found that CCT chaperonin binds free Cdc20 (Fig. 3C, lane 7). However, it does not bind free BubR1 (Fig. 3C, lane 9). In all these binding assays, there was no significant nonspecific binding to IgG beads (Fig. 3C). Because the chaperonin does not bind free BubR1, it is possible that it also does not bind to BubR1 in checkpoint complexes. That leaves the possibility that CCT chaperonin may bind to some available site(s) in the Cdc20 moiety of the checkpoint complexes (Discussion).
In their action in protein folding, chaperonins bind protein substrates in the absence of ATP, whereas their release is accompanied by the hydrolysis of ATP (17–19). We found that in this case, too, the dissociation of the chaperonin from mitotic checkpoint complexes requires ATP, as assayed by the release of CCT5 from immunoprecipitated complexes to supernatants (Fig. 3D). This finding suggests that the mechanisms of CCT-mediated protein folding and of MCC disassembly may by similar (Discussion).
Discussion
We have been studying the molecular mechanisms of the disassembly of MCC, a process required for the termination of the checkpoint-arrested state of APC/C. Evidence showed that there are several distinct pathways for MCC dissociation. Mad2 is released by the joint action of the TRIP13 AAA-ATPase and the Mad2-binding protein p31comet (11), whereas a part of Cdc20 is released from MCC by Cdk-catalyzed phosphorylation (16). We observed, however, that another part of the release of Cdc20 was phosphorylation independent, but ATP dependent (16). The present study was initiated to elucidate the latter process.
Using an assay to monitor the disassembly of recombinant checkpoint complexes, we confirmed that a considerable part of the release of Cdc20 from MCC is carried out by a phosphorylation-independent, but ATP-dependent process and furthermore found that a similar process is involved in the disassembly of the subcomplexes BC-1 and BC-2 (Fig. 1A). This process led to the partial purification of an ATP-dependent disassembly factor by anion exchange chromatography (Fig. 1B). Immunoblotting of the resolved fractions showed coincidence of disassembly activity with a subunit of the CCT/TRiC chaperonin, but not with some other chaperone proteins or ATPases (Fig. S2). Purification of the disassembly factor to near homogeneity by ATP affinity chromatography, followed by mass spectrometric analysis of tryptic peptides, identified all major proteins as subunits of the CCT/TRiC chaperonin (Fig. 2B). This identification was confirmed by the observation that recombinant CCT5 complex has activity in MCC dissociation (Fig. 3A).
It was very surprising to find a role of CCT chaperonin in MCC dissociation because to the best of our knowledge, this is a unique description of its involvement in the disassembly of an oligomeric complex. A great body of previous work described actions of CCT/TRiC chaperonin in the folding of proteins to their native states (reviewed in refs. 17–19). It is a complex composed of two rings of eight homologous subunits surrounding a central cavity, where folding is thought to take place. Initially, CCT was thought to have a specific role in the folding of the cytoskeletal proteins actin and tubulin, but subsequently it was found to have a much more general role in cellular protein folding. Thus, it was estimated that CCT/TRiC chaperonin is involved in the folding of up to 10–15% of newly synthesized proteins in animal cells (25). It was also reported to be involved in the folding of physiologically important substrates such as the cell cycle regulators Cdc20 (24) and Plk1 (26), tumor suppressors p53 and VHL, and Gs subunits of signaling complexes (reviewed in ref. 19). CCT/TRiC chaperonins do not recognize specific amino acid sequence motifs in substrates, but appear to have a broad binding specificity to structures such as hydrophobic surfaces (27). Proteins with high β-sheet propensity and WD-repeat containing proteins are preferred substrates for CCT/TRiC chaperonin. Thus, CCT chaperonin was shown to bind to the WD-repeat domain of Cdc20 in yeast (24).
At present, we can only speculate about the details of the molecular mechanisms by which CCT chaperonin disassembles mitotic checkpoint complexes. As mentioned above, to our knowledge this is a unique case in which CCT chaperonin is involved in complex disassembly. We note, however, that it was reported that CCT/TRiC chaperonin preferentially binds oligomeric proteins (27). Although this report was interpreted as being due to its role in the folding of a subunit to conformation suitable for complex assembly, binding could also be involved in the disassembly of some of these complexes. Concerning the moiety in mitotic checkpoint complexes recognized by the chaperonin, we observed that it binds not only MCC, but also the subcomplexes BC-1 and BC-2 (Fig. 3C). Because free BubR1 is not bound to the chaperonin (Fig. 3C), that leaves the Cdc20 moiety of the subcomplexes as a possible candidate for chaperonin binding. However, in MCC, and possibly also in BC-1 derived from MCC, a region in the WD-repeat domain of Cdc20 is already bound to BubR1 (28). In BC-2, the internal binding site of BubR1 is also bound to some part of the WD40-repeat domain of Cdc20, but the exact mode of this interaction is not known (29). If chaperonin binds a specific region of the Cdc20 moiety of mitotic checkpoint complexes, this may be in a part not occupied by BubR1 in either BC-1 or BC-2. However, it is also possible that CCT chaperonin does not bind to a specific region, but to any exposed hydrophobic region of Cdc20. Further investigation is required to clarify this problem.
After checkpoint complexes bind to CCT chaperonin in the absence of ATP, Cdc20 is released from BubR1, and BubR1 is released from chaperonin in a process that requires ATP hydrolysis (Fig. 3 B and D). This process is similar to events that take place in the binding and release of substrates for folding (17–19), but we do not know the intermediary processes in mitotic complex disassembly. It is generally assumed that the central cavity of the chaperonin complex is the site of protein folding. This chamber can accommodate proteins up to a size of ∼70 kDa. The folding of larger proteins may occur in a process in which only a domain enters and folds in the inner chamber (30). It is possible, for example, that the entry into this chamber of only a part of the mitotic checkpoint complex may initiate a conformational change in Cdc20 that leads to its dissociation from BubR1. Much more work is required to elucidate the intermediary events in the disassembly of mitotic checkpoint complexes by the CCT/TRiC chaperonin.
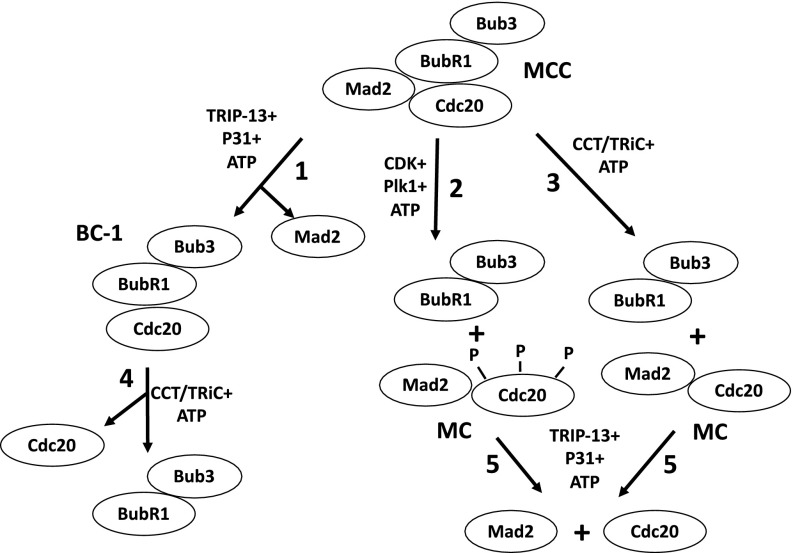
The identification of the CCT chaperonin-promoted dissociation process described in this paper fills a gap in our understanding of the processes by which free MCC is completely disassembled into its basic components. These processes are summarized in Fig. 4. Initially, MCC may be subjected to disassembly by three pathways: Mad2 is released by the joint action of TRIP13 and p31comet (Fig. 4, step 1), whereas Cdc20 is dissociated by phosphorylation (step 2) and by the action of CCT/TRiC chaperonin (step 3). The subsequent disassembly of the subcomplexes released by the above processes is then completed by the complementing activities: BC-1 is further dissociated by CCT/TRiC chaperonin, as shown in this paper (step 4), whereas the subcomplex Mad–Cdc20 is subjected to disassembly by TRIP13 and p31comet (ref. 11 and step 5). As discussed above, the complete disassembly of both pools of free MCC and of APC/C-bound MCC is required for exit from the checkpoint-arrested state. Although we have now detailed knowledge of the pathways of the disassembly of free MCC, the mechanisms of the release and disassembly of APC/C-bound MCC remain a challenge for future investigations.
Fig. 4.
Pathways of disassembly of MCC. See the text. BC-1, Cdc20 bound to the N-terminal binding site of BubR1 (15); MC, Mad2 bound to Cdc20. Bub3 is constitutively bound to BubR1 throughout the cell cycle.
Materials and Methods
Preparations.
Recombinant MCC and BC-2 were formed by coinfection of SF9 insect cells with baculoviruses expressing the appropriate proteins (streptavidin-binding peptide–BubR1, Myc–Cdc20, and his6–Bub3 with or without flag–Mad2, followed by sequential purification steps as described previously (15). BC-1 subcomplex was generated by subjecting recombinant MCC to joint action of TRIP13 and p31comet as described (15). Extracts of nocodazole-arrested HeLa cells were prepared as described previously (31). CCT/TRiC chaperonin was purified from extracts of HeLa cells as described in legends to Figs. 1B and 2A and Fig. S3. Protein kinase Cdk1–GST–Δ88 cyclin B (referred to as “Cdk1–cyclin B”) was prepared and purified as described (32). The Xenopus homolog of Plk1 and its catalytically inactive N172A mutant were prepared as previously (33).
Immunoprecipitation and Immunoblotting.
Rabbit polyclonal antibodies directed against human proteins were used for immunoprecipitation. All antibodies were purified by affinity chromatography on their respective antigens: α-Cdc27, 17-amino acid C-terminal peptide of Cdc27; α-BubR1, a his6-tagged 38- to 468-aa fragment of BubR1; and α-Cdc20, the corresponding his6-tagged full-length human protein. For immunoprecipitation, all antibodies were bound to Affi-Prep Protein A beads (Bio-Rad) at a concentration of 0.5 mg/mL of packed beads. For sham immunoprecipitations, purified rabbit IgG (Pierce) was bound to the Protein A beads at a similar concentration. For immunoblotting of human proteins, the following monoclonal antibodies were used: BubR1 (BD Transduction Laboratories 612053); Cdc20 (Santa Cruz Biotechnology sc-13162); Mad2 (MBL Laboratories K0167); c-Myc (Cell Signaling 2276S); TCP-1α/CCT1 (Santa Cruz Biotechnology sc-53454); TCP-1ε/CCT5 (Santa Cruz Biotechnology sc-374554; HSP70 (Cell Signaling 4872S); HSP90 (Santa Cruz Biotechnology sc-69703); and VCP/p97 (Santa Cruz Biotechnology sc-20799). Immunoblots were detected and quantified with fluorescently labeled secondary antibodies using Odyssey (Li-Cor) scanner.
Assay of Disassembly of Mitotic Checkpoint Complexes.
Recombinant MCC, BC-1, or BC-2 (50 pmol of protein, all containing myc–Cdc20) were bound to 20 μL anti-BubR1 beads by absorption at 4 °C with gentle agitation for 2 h, followed by washing twice with a buffer consisting of 50 mM Tris⋅HCl (pH 7.2), 20% (vol/vol) glycerol, 1 mg/mL BSA, 1 mM DTT, and 0.3 M NaCl, and then twice with the same buffer without NaCl. Beads were incubated in a volume of 30 µL with a reaction mixture that contained 50 mM Tris⋅HCl (pH 7.6), 5 mM MgCl2, 1 mM DTT, and 10% (vol/vol) glycerol, and 2 mM ATP and enzyme source as indicated. When ATP was omitted, hexokinase, 0.4 mg/mL and 2-deoxyglucose, 10 mM were also added. Following incubation at 30 °C with shaking at 1,400 rpm for 1 h, samples were transferred to ice, centrifuged, and the supernatants were passed through 0.45-μM centrifugal filters to remove residual beads. The release of myc–Cdc20 from anti–BubR1-bound recombinant mitotic complexes to supernatants was determined by immunoblotting with anti-myc antibody. Control experiments showed that BubR1 bound to anti-BubR1 beads was not released (Fig. 1B), thus the release of Cdc20 or Mad2 to supernatants represented dissociation from BubR1.
Acknowledgments
We thank Dr. Jonathan A. King (Massachusetts Institute of Technology) for generously providing the CCT5 expression vector. This work was supported by grants from the Israel Science Foundation and the Israel Cancer Research Fund. A part of the work was performed at the Marine Biological Laboratory, supported by the Ricbac Foundation for Research in Cell Biology and Cancer.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620451114/-/DCSupplemental.
References
- 1.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 2.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012;22(22):R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Jia L, Kim S, Yu H. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem Sci. 2013;38(6):302–311. doi: 10.1016/j.tibs.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Chang L, Barford D. Insights into the anaphase-promoting complex: A molecular machine that regulates mitosis. Curr Opin Struct Biol. 2014;29:1–9. doi: 10.1016/j.sbi.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Sivakumar S, Gorbsky GJ. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat Rev Mol Cell Biol. 2015;16(2):82–94. doi: 10.1038/nrm3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eytan E, Sitry-Shevah D, Teichner A, Hershko A. Roles of different pools of the mitotic checkpoint complex and the mechanisms of their disassembly. Proc Natl Acad Sci USA. 2013;110(26):10568–10573. doi: 10.1073/pnas.1308928110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446(7138):921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 8.Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10(12):1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansfeld J, Collin P, Collins MO, Choudhary JS, Pines J. APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nat Cell Biol. 2011;13(10):1234–1243. doi: 10.1038/ncb2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miniowitz-Shemtov S, Teichner A, Sitry-Shevah D, Hershko A. ATP is required for the release of the anaphase-promoting complex/cyclosome from inhibition by the mitotic checkpoint. Proc Natl Acad Sci USA. 2010;107(12):5351–5356. doi: 10.1073/pnas.1001875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eytan E, et al. Disassembly of mitotic checkpoint complexes by the joint action of the AAA-ATPase TRIP13 and p31(comet) Proc Natl Acad Sci USA. 2014;111(33):12019–12024. doi: 10.1073/pnas.1412901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, et al. Thyroid hormone receptor interacting protein 13 (TRIP13) AAA-ATPase is a novel mitotic checkpoint-silencing protein. J Biol Chem. 2014;289(34):23928–23937. doi: 10.1074/jbc.M114.585315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miniowitz-Shemtov S, Eytan E, Kaisari S, Sitry-Shevah D, Hershko A. Mode of interaction of TRIP13 AAA-ATPase with the Mad2-binding protein p31comet and with mitotic checkpoint complexes. Proc Natl Acad Sci USA. 2015;112(37):11536–11540. doi: 10.1073/pnas.1515358112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye Q, et al. TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. eLife. 2015;4:4. doi: 10.7554/eLife.07367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaisari S, Sitry-Shevah D, Miniowitz-Shemtov S, Hershko A. Intermediates in the assembly of mitotic checkpoint complexes and their role in the regulation of the anaphase-promoting complex. Proc Natl Acad Sci USA. 2016;113(4):966–971. doi: 10.1073/pnas.1524551113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miniowitz-Shemtov S, et al. Role of phosphorylation of Cdc20 in p31(comet)-stimulated disassembly of the mitotic checkpoint complex. Proc Natl Acad Sci USA. 2012;109(21):8056–8060. doi: 10.1073/pnas.1204081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: Physiology and mechanism. Annu Rev Cell Dev Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- 18.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 19.Lopez T, Dalton K, Frydman J. The mechanism and function of group II chaperonins. J Mol Biol. 2015;427(18):2919–2930. doi: 10.1016/j.jmb.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26(1):127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 21.Lénárt P, et al. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17(4):304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 22.Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6(7):519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 23.Sergeeva OA, et al. Human CCT4 and CCT5 chaperonin subunits expressed in Escherichia coli form biologically active homo-oligomers. J Biol Chem. 2013;288(24):17734–17744. doi: 10.1074/jbc.M112.443929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camasses A, Bogdanova A, Shevchenko A, Zachariae W. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol Cell. 2003;12(1):87–100. doi: 10.1016/s1097-2765(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 25.Thulasiraman V, Yang CF, Frydman J. In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J. 1999;18(1):85–95. doi: 10.1093/emboj/18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, et al. CCT chaperonin complex is required for the biogenesis of functional Plk1. Mol Cell Biol. 2005;25(12):4993–5010. doi: 10.1128/MCB.25.12.4993-5010.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yam AY, et al. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol. 2008;15(12):1255–1262. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484(7393):208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 29.Diaz-Martinez LA, et al. The Cdc20-binding Phe box of the spindle checkpoint protein BubR1 maintains the mitotic checkpoint complex during mitosis. J Biol Chem. 2015;290(4):2431–2443. doi: 10.1074/jbc.M114.616490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rüßmann F, et al. Folding of large multidomain proteins by partial encapsulation in the chaperonin TRiC/CCT. Proc Natl Acad Sci USA. 2012;109(52):21208–21215. doi: 10.1073/pnas.1218836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braunstein I, Miniowitz S, Moshe Y, Hershko A. Inhibitory factors associated with anaphase-promoting complex/cylosome in mitotic checkpoint. Proc Natl Acad Sci USA. 2007;104(12):4870–4875. doi: 10.1073/pnas.0700523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golan A, Yudkovsky Y, Hershko A. The cyclin-ubiquitin ligase activity of cyclosome/APC is jointly activated by protein kinases Cdk1-cyclin B and Plk. J Biol Chem. 2002;277(18):15552–15557. doi: 10.1074/jbc.M111476200. [DOI] [PubMed] [Google Scholar]
- 33.Moshe Y, Boulaire J, Pagano M, Hershko A. Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc Natl Acad Sci USA. 2004;101(21):7937–7942. doi: 10.1073/pnas.0402442101. [DOI] [PMC free article] [PubMed] [Google Scholar]