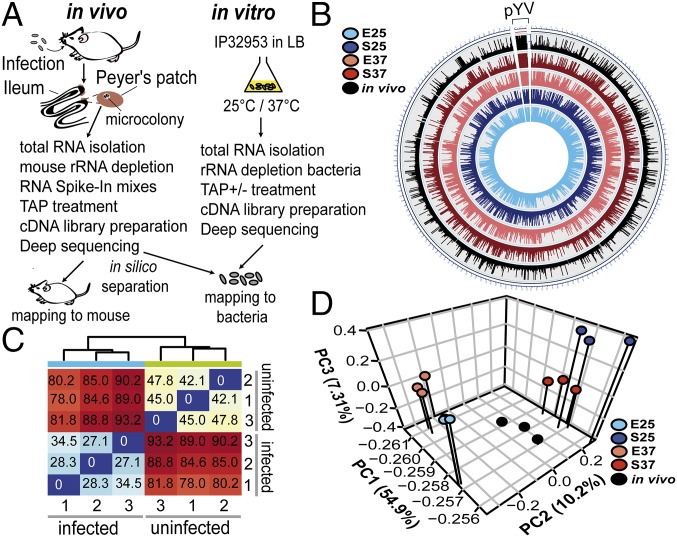
Fig. 1.
Tissue dual RNA-seq workflow and global reports. (A) For tissue dual RNA-seq, female BALB/c mice were orally infected with 2 × 108 cfu Y. pseudotuberculosis IP32953 (infected) or 1× PBS (uninfected). For in vitro RNA-seq, IP32953 was grown in LB to exponential or stationary phase at 25 °C or 37 °C. Total RNA was isolated from Peyer’s patches of mice or bacterial cultures processed for preparation of strand-specific barcoded cDNA libraries and sequenced (SI Appendix). cDNA reads were separated in silico by mapping to the mm10 and the IP32953 genomes. TAP, tobacco acid pyrophosphatase. (B) Circos plot visualizing reads per kilobase transcript length per million mapped reads-normalized expression values of in vitro and in vivo RNA-seq data for the IP32953 chromosome (NC_006155.1) and pYV virulence plasmid (NC_006153.2). E25, exponential phase 25 °C; E37, exponential phase 37 °C; in vivo, infected Peyer’s patches; S25, stationary phase 25 °C; S37, stationary phase 37 °C. (C) Heat map of Euclidian sample distances of rlog-transformed read counts for triplicate mouse RNA-seq data from RNA pools of uninfected and infected Peyer’s patches. (D) Principal component (PC) analysis of mean centered and scaled rlog-transformed read count values of bacterial in vitro and in vivo RNA-seq data for the Y. pseudotuberculosis IP32953 core genome and the pYV virulence plasmid.