Significance
To study mechanisms of interspecies prion transmission, we used transgenic mice that recapitulate natural disease susceptibilities. Predictably, prions from a particular species inefficiently caused disease in mice expressing prion proteins from different species. However, in contrast to the expected adaptation that allows efficient prion replication in the new host, we discovered a process in which pathogenic prions, despite causing disease on primary passage, failed to adapt for sustained propagation in the new host, instead preserving the strain and host range characteristics of the original prions. Our findings provide insights into how prions replicate during species transitions, help clarify unresolved epidemiological features of prion disorders, and have considerable bearing on future disease risk assessments.
Keywords: prion strains, conformational selection, transgenic mice, species barriers, adaptation
Abstract
Adaptation of prions to new species is thought to reflect the capacity of the host-encoded cellular form of the prion protein (PrPC) to selectively propagate optimized prion conformations from larger ensembles generated in the species of origin. Here we describe an alternate replicative process, termed nonadaptive prion amplification (NAPA), in which dominant conformers bypass this requirement during particular interspecies transmissions. To model susceptibility of horses to prions, we produced transgenic (Tg) mice expressing cognate PrPC. Although disease transmission to only a subset of infected TgEq indicated a significant barrier to EqPrPC conversion, the resulting horse prions unexpectedly failed to cause disease upon further passage to TgEq. TgD expressing deer PrPC was similarly refractory to deer prions from diseased TgD infected with mink prions. In both cases, the resulting prions transmitted to mice expressing PrPC from the species of prion origin, demonstrating that transmission barrier eradication of the originating prions was ephemeral and adaptation superficial in TgEq and TgD. Horse prions produced in vitro by protein misfolding cyclic amplification of mouse prions using horse PrPC also failed to infect TgEq but retained tropism for wild-type mice. Concordant patterns of neuropathology and prion deposition in susceptible mice infected with NAPA prions and the corresponding prion of origin confirmed preservation of strain properties. The comparable responses of both prion types to guanidine hydrochloride denaturation indicated this occurs because NAPA precludes selection of novel prion conformations. Our findings provide insights into mechanisms regulating interspecies prion transmission and a framework to reconcile puzzling epidemiological features of certain prion disorders.
Sheep scrapie, bovine spongiform encephalopathy (BSE), bovine amyloidotic spongiform encephalopathy (BASE), chronic wasting disease (CWD) of cervids, transmissible mink encephalopathy (TME), and human Creuzfeldt–Jakob disease (CJD) are fatal neurodegenerative disorders caused by prions, which are transmissible proteinaceous agents lacking nucleic acids. Replication involves corruption of the host-encoded cellular form of the prion protein (PrPC) by its abnormally conformed infective counterpart, referred to as PrPSc (1). Inoculation of prions into individuals of the same species typically causes disease with reproducible clinical signs and uniformly consistent incubation periods. Cross-species transmission is less efficient, and characterization requires disease comparisons during primary and secondary transmissions to the new host (reviewed in ref. 2). On first passage, all inoculated animals either remain asymptomatic for extended periods (3–9) or may develop disease after a prolonged asymptomatic phase (8, 10, 11). In a third alternative, only a subpopulation develops disease, with variable incubation times (8, 12, 13). However, in all cases, secondary transmission results in shorter, synchronous times to disease in all recipients. Collectively, these properties are indicative of relatively inefficient interspecies transmission, albeit to varying degrees, and subsequent adaptation for optimal and stable replication in the new host.
The importance of primary structural incompatibilities between PrPSc constituting the prion and PrPC expressed in the new host paved the way for transgenic (Tg) models that abrogate transmission barriers to prions from various species in mice (14) or that model susceptibility of at-risk or seemingly resistant species (15, 16). Prion strain properties also influence interspecies prion transmissions. Heritable strain properties, including incubation times and neuropathological profiles, are enciphered within distinct PrPSc conformations (17, 18). Interspecies prion transmission often results in the establishment of strains that are distinct from the original (11, 19, 20), with concomitant changes in PrPSc conformation (20). The conformational selection model reconciled the effects of primary and higher order PrP structures on adaptive transmission by postulating that PrPC in the new host selectively propagates species-optimized prions from an ensemble of quasi-species conformations produced during replication in the ancestral host (21).
Recent studies focused on the role of specific structural elements during adaptive transspecies transmissions, particularly the loop connecting PrP β2 and α2 regions (22, 23), which is disordered in most species but conformationally defined in others such as elk and bank vole PrPC. Although β2–α2 loop rigidity was thought to potentiate PrPC to PrPSc conversion (24), subsequent studies revealed that PrPC of species considered resistant to prions, including horses, also contained rigid β2–α2 loops (25–27). Adding additional complexity, Tg mice expressing mouse PrPC containing rigid β2–α2 loops from elk or horse PrP spontaneously developed prion diseases (28, 29). To clarify the effect of the horse PrP β2–α2 loop on prion conversion and to model susceptibility of this at-risk species, we produced Tg mice expressing horse PrP. These TgEq mice were resistant to disease following inoculation, with the exception of SSBP/1 scrapie prions, which produced disease in a subset of TgEq. Surprisingly, the resulting horse prions failed to cause disease upon transmission to additional TgEq but paradoxically retained the strain and host range properties of the ancestral prions. Here we characterize several examples and address the mechanistic basis of this unusual replicative phenomenon.
Results
Addressing the Capacity of Horse PrPC to Propagate Naturally Occurring Prions.
Spontaneous neurodegeneration in Tg mice in which the mouse PrP β2–α2 loop was replaced by that of horse PrP (29) contrasted structural predictions of intrinsic horse PrPC resistance to conformational conversion (27). To address this inconsistency, we created TgEq expressing horse PrPC (EqPrPC) at levels approximately twofold higher than PrPC in horse brains (Fig. S1). Aged TgEq and mice inoculated with brain extracts from uninfected animals remained free of disease (Table 1, primary passage of prions to TgEq), indicating that EqPrPC expression in TgEq does not result in spontaneous disease. Disease and PrPSc accumulation were registered after 454 and 481 d in two of six TgEq inoculated intracerebrally (ic) with SSBP/1 scrapie prions (Table 1, primary passage of prions to TgEq, Fig. 1A, and Fig. S2), whereas the remaining inoculated TgEq remained free of disease and PrPSc accumulation for 600 d (Fig. 1A). We refer to these horse prions as Eq-SSBP/1. TgEq were entirely resistant to ic inoculation with elk or deer CWD; BSE or BASE; TME; atypical scrapie; and the mouse-adapted scrapie isolate referred to as RML (Rocky Mountain Laboratory) (Table 1, primary passage of prions to TgEq). Our results establish that, despite a general resistance to conformational conversion, EqPrPC is capable of supporting pathogenic horse prion replication.
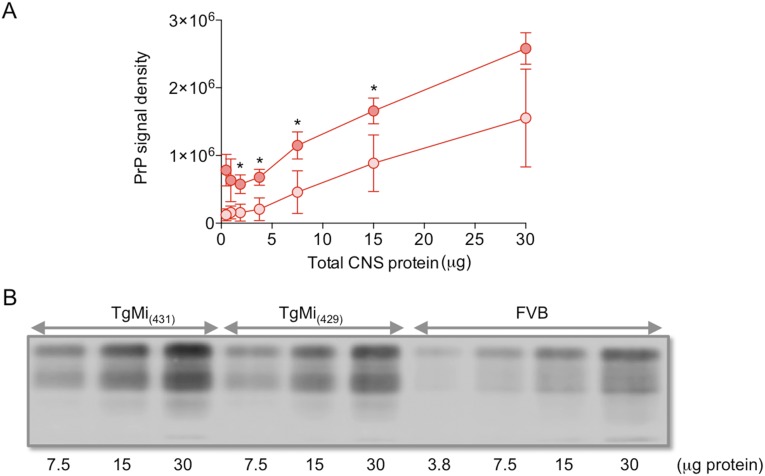
Fig. S1.
PrP expression in the central nervous system of TgEq and TgMi mice. The amount of total protein in brain extracts of animals was determined using BCA, and homogenates containing various indicated amounts of protein were analyzed by SDS/PAGE, followed by Western blotting. Membranes were probed with mAb PRC5 or 6H4 overnight at 4 °C, followed by IRDye 800 CW goat anti-mouse IgG secondary antibody (LI-COR Biosciences) for 1 h. Membranes were scanned with the Odyssey CLx infrared imaging system (LI-COR Biosciences), and signals were densitometrically analyzed with Odyssey CLx Image Studio software (LI-COR Biosciences). (A) Mean densitometric values of PrP-specific signals were calculated following analysis of whole brain extracts from four TgEq and the cerebral cortices of four horse brains. Values were plotted against amounts of total protein loaded. Darker red circles, TgEq; lighter red circles, horse. Error bars represent the SEM. Asterisks (*) indicate differences in the levels of horse PrP expression between TgEq and horse. The level of horse PrP expression in whole brains from TgEq was estimated to be ∼twofold higher than the levels in the cerebral cortices of horse brain. (B) Western blots showing expression levels of PrPC in the brains of wild-type FVB mice and two lines of Tg mice expressing mink PrPC, referred to as TgMi(431) and TgMi(429). All Tg mice were maintained on an FVB/Prnp0/0 background in which endogenous mouse PrPC is not expressed.
Table 1.
NAPA
| Inoculum | TgEq | TgOvV | TgOvA | C57BL/6 | TgD | TgMi(F429) | TgMi(F431) |
| Primary passage of prions to TgEq | |||||||
| None | >547 (0/7) | ||||||
| Cattle NBH | >872 (0/9) | ||||||
| Horse NBH | >810 (0/6) | ||||||
| Elk CWD | >591 (0/6) | ||||||
| Deer CWD | >607 (0/7) | ||||||
| BSE | >833 (0/12) | ||||||
| BASE | >813 (0/11) | ||||||
| RML | >586 (0/7) | 140 ± 2 (5/5) | |||||
| TME | >624 (0/6) | ||||||
| Atypical scrapie | >810 (0/12) | ||||||
| SSBP/1 (p1)* | 468 ± 14 (2/6) | 132 ± 2 (8/8) | 412 ± 49 (6/6) | ||||
| Secondary passage of NAPA horse prions | |||||||
| Horse p2† | >500 (0/6) | 162 ± 2 (4/4) | 431 ± 20 (4/4) | ||||
| PMCA-Eq-SSBP/1 | >503 (0/7) | ||||||
| PMCA-Eq-BSE | >813 (0/20) | ||||||
| PMCA-Eq-RML | >524 (0/8) | 246 ± 17 (5/5) | |||||
| Primary passage of TME prions | |||||||
| Cloned (p1) | 572 ± 1 (6/6) | 130 ± 4 (6/6) | 109 ± 2 (8/8) | ||||
| Cloned (p1) | 575 ± 7 (6/6)‡ | 128 ± 2 (7/7) | 123 ± 3 (8/8) | ||||
| Uncloned | 279 ± 15 (6/6) | 262 ± 17 (7/7) | |||||
| Serial passage of NAPA deer prions | |||||||
| Deer p2§ | >580 (0/8)‡ | 167 ± 10 (6/6) | 153 ± 9 (6/6) | ||||
| Deer p3¶ | 236 ± 55 (4/8) | 247 (1/8) | |||||
Incubation times are expressed as the mean time to disease onset in days (d), ± the standard error of the mean (SEM). In each case the number of diseased mice/number inoculated, excluding mice dying from intercurrent illnesses, is shown. Numbers in italics indicate transmissions that produced no disease. NBH, normal brain homogenate.
Transmissions to TgOv mice, previously reported (38).
Horse p2 refers to second passage of horse prions from the CNS of diseased TgE resulting from inoculation with SSBP/1 during p1.
Source of material for subsequent transmission(s).
Deer p2 refers to passage of deer prions from the CNS of diseased TgD resulting from inoculation with TME during p1.
Deer p3 refers to passage of deer prions from the CNS of an asymptomatic TgD following p2.
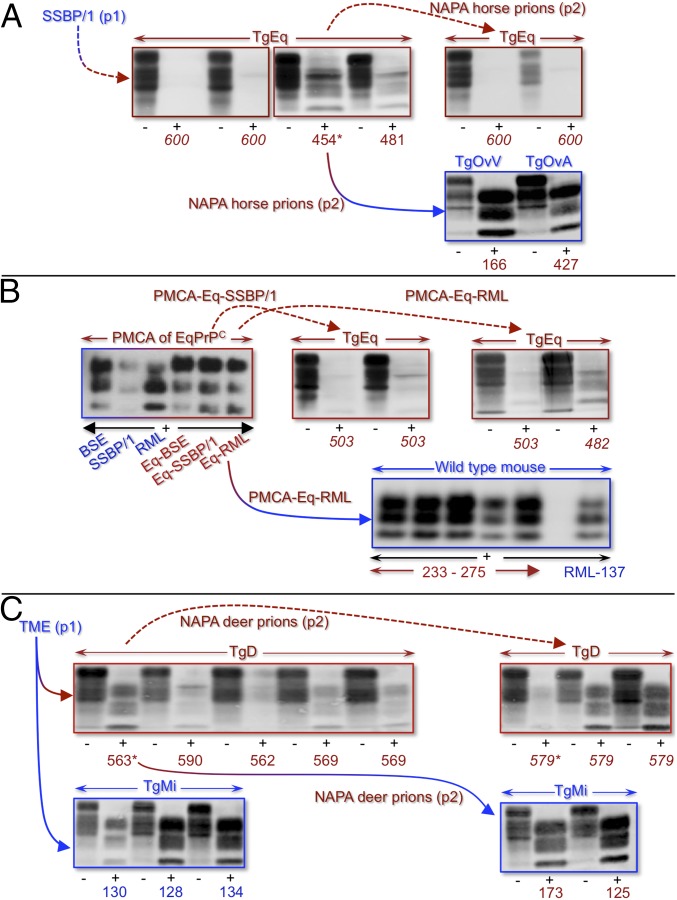
Fig. 1.
Western blotting of PrPSc in Tg mouse brains during NAPA. Species of prion origin and adaptation are indicated in blue and red. Solid lines with transitional colors, productive interspecies transmissions; solid curved arrows, disease transmission to all recipients; broken curved arrows, incomplete or no disease transmission; asterisk (*), animals used in transmissions; italicized numbers, asymptomatic mice; nonitalicized numbers, symptomatic mice. Samples treated (+) or not (–) with PK, and times of sacrifice (days) are shown beneath lanes. (A) NAPA horse prions in TgEq. (Upper Left) PrPSc in two diseased mice following primary passage (p1) of SSBP/1 to TgEq; no PrPSc in asymptomatic TgEq. (Upper Right) No PrPSc in asymptomatic TgEq following p2 of NAPA horse prions. (Lower) PrPSc in diseased TgOvV and TgOvA following p2 of NAPA horse prions. (B) NAPA horse prions produced by sPMCA of BSE, SSBP/1, and RML using PrPC from horse brain. (Top Left) PrPSc in unamplified BSE, SSBP/1, and RML samples (blue); corresponding NAPA horse prions after 20 rounds of sPMCA (red). (Top Middle) Asymptomatic TgEq inoculated with PMCA-Eq-SSBP/1. (Top Right) Asymptomatic TgEq inoculated with PMCA-Eq-RML. (Bottom) Diseased C57BL/6 mice inoculated with PMCA-Eq-RML. RML-137, RML infected wild-type mouse with prion disease after 137 d. (C) NAPA deer prions in TgD. (Top Left) PrPSc accumulation in diseased TgD after 560 d following p1 of TME. (Top Right) PrPSc in asymptomatic TgD after ∼580 following p2 of NAPA deer prions. 579*, asymptomatic TgD killed after 579 d, used in p3 of NAPA deer prions to TgMi (Table 1, primary passage of TME prions). (Lower Right) PrPSc in diseased TgMi(F431) following p2 of NAPA deer prions. (Lower Left) PrPSc in diseased TgMi(F431) following p1 of TME.
Fig. S2.
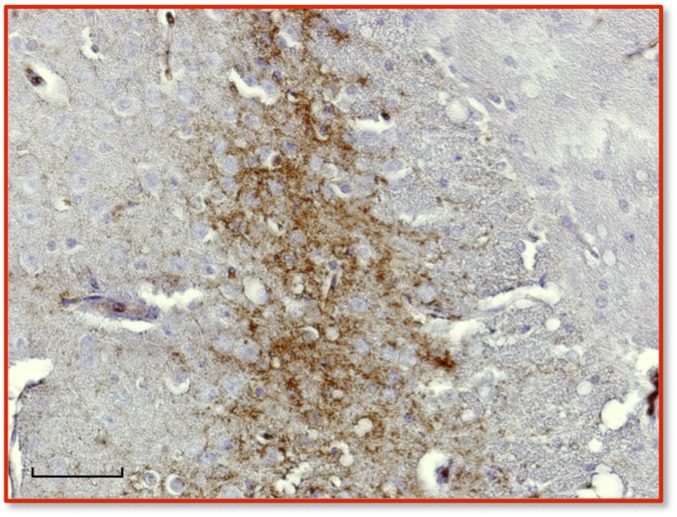
Immunohistochemical analysis of PrPSc in the central nervous system of diseased TgEq mice infected with SSBP/1. Immunohistochemical (IHC) analysis was performed as previously described (14) from sections of cortex taken at the region of the septum using mAb 6H4 as primary antibody and IgG1 biotinylated goat anti-mouse as secondary antibody (SouthernBiotech). Digitized images were obtained by light microscopy at 40× magnification using a Nikon Eclipse E600 microscope equipped with a Nikon DMX 1200F digital camera. (Scale bar, 50 μm.)
Transient Prion Adaptation During Interspecies Transmission.
Assessment of prion adaptation requires comparison of primary and subsequent prion transmissions to a new host, with species-adapted prions producing shorter, synchronous times to disease in all inoculated recipients on secondary passage. The absence of disease and PrPSc accumulation upon serial transmission of Eq-SSBP/1 to additional TgEq and their efficient transmission to TgOv mice were therefore unexpected (horse p2 in Table 1, secondary passage of NAPA horse prions, and Fig. 1A). The mean incubation time of horse prions was equivalent to that of ancestral SSBP/1 in TgOvA (P = 0.78) and 25% longer than SSBP/1 in TgOvV (P < 0.0001). TgOvA and TgOvV express sheep PrP with either alanine (A) or valine (V) at residue 136 at levels approximately equal to PrPC in mouse brains (30). We conclude that while disease in SSBP/1-infected TgEq resulted from replication of an EqPrPC-compatible prion conformer, the ensuing pathogenic horse prions were not permanently adapted for EqPrPC conversion, but instead retained disease potential in animals expressing PrPC from the species of prion origination. We refer to this process as nonadaptive prion amplification (NAPA).
NAPA in Other Settings.
To address whether these unconventional properties resulted from a peculiar resistance of EqPrPC to prion conversion, we sought additional examples of NAPA. When we challenged CWD-susceptible Tg mice (TgD) expressing deer PrP at levels ∼fivefold higher than PrPC in mouse brain (3), with two cloned preparations of TME (31), all mice developed disease and accumulated PrPSc after ∼575 d (Table 1, primary passage of TME prions, and Fig. 1C). Although this protracted incubation time was consistent with a transmission barrier, the complete attack rate and invariant incubation times suggested that adaptation of TME might have occurred in diseased TgD. However, serial transmission of these pathogenic deer prions, referred to as D-TME, to additional TgD failed to produce disease within ∼580 d (Table 1, serial passage of NAPA deer prions), despite PrPSc accumulation (p2 in Fig. 1C). To address whether D-TME prions instead retained the ability to efficiently convert mink PrPC (MiPrPC) and consequently were propagated by NAPA, we produced Tg mice expressing MiPrPC (TgMi). Two lines, TgMi(F429) and TgMi(F431), expressed MiPrP at levels approximately equal to PrP in the brains of wild-type mice (Fig. S1B). In accordance with previous studies (32), the barrier to TME transmission in mice was eliminated by MiPrP expression in TgMi. Mean incubation times of cloned TME (31) in TgMi(F429) and TgMi(F431) were ∼130 and ∼115 d, whereas that of uncloned TME was twofold longer (Table 1, primary passage of TME prions). D-TME prions produced disease and PrPSc upon serial transmission to TgMi(F429) and TgMi(F431) after ∼160 d (Table 1, serial passage of NAPA deer prions, and Fig. 1C). Our results indicate that TME prions also replicate without adapting for optimal propagation in TgD. Brain extracts of an asymptomatic, PrPSc-positive TgD infected with D-TME killed at ∼580 d during p2 also produced disease and PrPSc in 50% of inoculated TgMi(F429) after ∼235 d and a single TgMi(F431) (Table 1, serial passage of NAPA deer prions), suggesting that NAPA may result in the asymptomatic accumulation of prions, which is similar to previous findings during infection of a nonpermissive host (33).
Protein misfolding cyclic amplification (PMCA) recapitulates PrPSc replication through a cyclical process involving incubation and sonication in vitro (34). In serial PMCA, prions replicate indefinitely through successive rounds of dilutions and amplification, and this strategy has been used to traverse species barriers and generate species-adapted prions (35). Because sPMCA generated infectious rabbit prions, a species that was considered resistant to prion disease (36), we used the same approach to produce horse prions. SSBP/1 converted EqPrPC from horse brain following 20 rounds of sPMCA, with EqPrPSc initially observed at round 14. These prions were designated PMCA-Eq-SSBP/1 (Fig. 1B). sPMCA also overcame the transmission barrier for BSE and RML prions that we observed in TgEq (Table 1, primary passage of prions to TgEq), producing PMCA-Eq-BSE and PMCA-Eq-RML prions (Fig. 1B). We challenged TgEq with Eq-SSBP/1, Eq-BSE, and Eq-RML horse prions produced following 20 rounds of sPMCA (Fig. 1B). Because sPMCA-derived rabbit prions caused disease in Tg mice expressing rabbit PrPC (36), we anticipated efficient disease induction in all cases. Nonetheless, all inoculated TgEq remained asymptomatic for >500 d (Table 1, secondary passage of NAPA horse prions) despite low-level accumulation of PrPSc. We next asked if PMCA-Eq-RML caused disease in the species of prion origin, in this case wild-type mice, and therefore replicated by NAPA. Disease and PrPSc accumulation was registered in all inoculated mice with a mean incubation time ∼75% longer than RML prions (P < 0.0001) (Table 1, secondary passage of NAPA horse prions, and Fig. 1B). We conclude that sPMCA recapitulates the features of NAPA observed in vivo. Moreover, unlike rabbits and Tg mice expressing rabbit PrPC, which are susceptible to sPMCA-adapted rabbit prions, sPMCA-derived horse prions are only transiently adapted for EqPrPC conversion and, like NAPA prions produced in vivo, retain pathogenic potential for their species of origin.
Comparing the Properties of Source and NAPA Prions.
The similar host range of NAPA prions and their ancestral counterparts suggested that strain properties of SSBP/1 and TME prions remained unchanged following replication in TgEq and TgD. We therefore compared the disease characteristics of TgOv infected with either SSBP/1 or NAPA horse prions and of TgMi infected with either TME or NAPA deer prions. Histoblotting revealed comparable PrPSc distributions in TgOvV infected with either NAPA horse prions or SSBP/1 (Fig. 2A). Distributions of PrPSc and neuropathology were also concordant in TgMi infected with NAPA deer prions or the mink prions from which they originated (Fig. 2 B and C).
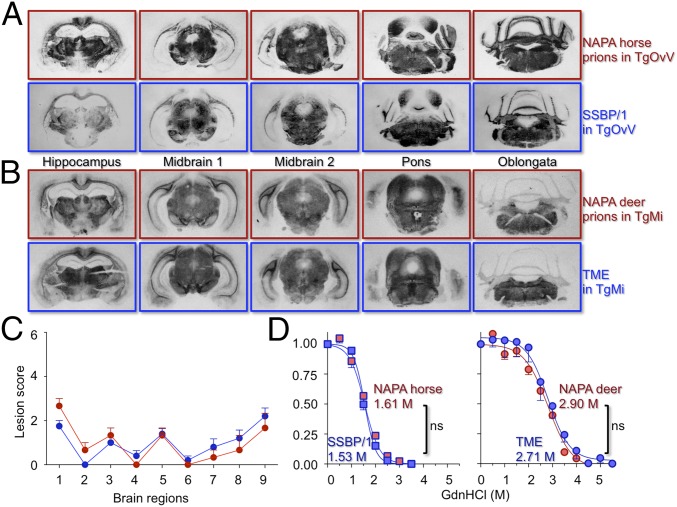
Fig. 2.
Neuropathology and PrPSc properties in representative mice infected with NAPA prions and ancestral counterparts. PrPSc distribution in PK-treated, mAb 6H4 probed histoblots of (A) TgOvV infected with SSBP/1 (blue panels) or NAPA horse prions (red panels). (B) TgMi infected with TME (blue panels) or NAPA deer prions (red panels). (C) Extent of vacuolar degeneration in hematoxylin and eosin-stained, paraffin-embedded sections of the following: 1, medulla; 2, cerebellum; 3, midbrain; 4, hypothalamus; 5, thalamus; 6, hippocampus; 7, paraterminal body; 8, cerebral cortex/hippocampus; and 9, cerebral cortex/septum. A vacuolation severity score of 0, for none, and 4 (maximum) was recorded. Error bars, average score per field ± SEM. TME-infected TgMiF431 brains (n = 5, blue circles) or NAPA deer prions (n = 3, red circles). (D) Percentage of protease-resistant PrPSc as a function of Gdn.HCl concentration. Fapp, fraction of apparent PrPSc = (maximum signal – individual signal)/(maximum signal – minimum signal). The sigmoidal dose–response curve was plotted using a four-parameter algorithm and nonlinear least-square fit. Statistical differences of the GdnHCl1/2 between matched, best-fitted curves were calculated. ns, not significant. Error bars, SEM from three animals per group. Prions from the species of origin, blue; counterpart NAPA prions, red. Shown are TgOvV infected with SSBP/1 (blue squares) or NAPA horse prions (red squares), and TgMi infected with TME (blue circles) or NAPA deer prions (red circles).
To assess the effects of NAPA on PrPSc stability, a biochemical property associated with prion conformation (20), which is the generally accepted basis of strain variability (17, 18), we performed guanidine hydrochloride (GdnHCl) denaturation profiling. Although the stabilities of TME and SSBP/1 PrPSc in TgMi and TgOvV were different (GdnHCl1/2 ∼2.8 and 1.55), there were no differences in the stabilities of MiPrPSc produced following infection of TgMi with TME or the corresponding NAPA deer prions (P = 0.44), nor between the stabilities of EqPrPSc produced following infection of TgEq with SSBP/1 or the corresponding NAPA horse prions (P = 0.09) (Fig. 2D). We conclude that, despite transient adaptation in mice expressing horse or deer PrPC, the strain properties of SSBP/1 and TME remained unchanged during NAPA.
Prion Replication During NAPA.
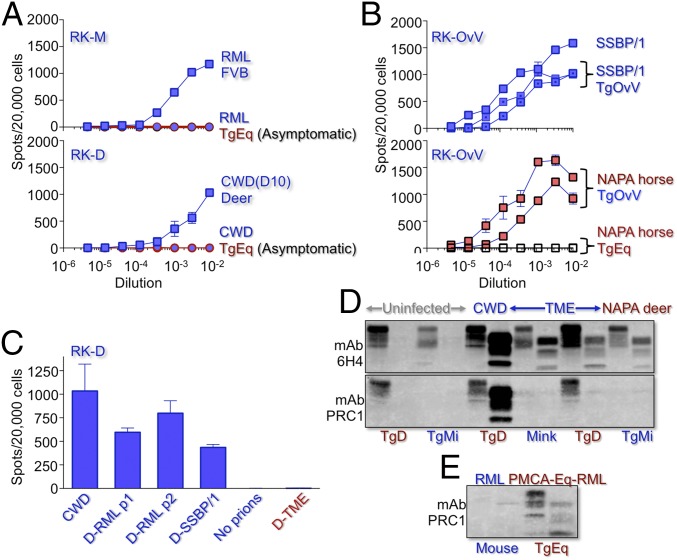
We took several measures to exclude the possibility that these unorthodox transmission profiles were related to the persistence of prions in primary inocula. In previous studies, rabbit kidney epithelial RK13 cells engineered to produce foreign PrPC supported prion replication from the corresponding species, allowing quantification of prion titers in a cell culture setting (37). We produced RK-13 cells expressing mouse PrPC (RK-M) or deer PrPC (RK-D) to similarly allow titration of mouse and deer CWD prions. Although RML and CWD prions replicated to high titers in the brains of inoculated FVB mice and deer, respectively (Fig. 3A), prions were undetectable in the brains of asymptomatic TgEq >500 d after challenge by RML or CWD (Fig. 3A), reflecting inoculum clearance in the absence of prion replication. To assess prion titers in TgOvV mice infected with SSBP/1 or Eq-SSBP/1, we produced RK-13 cells expressing ovine PrP with V at residue 136 (RK-OvV), which are functionally equivalent to scrapie-susceptible Rov9 cells (38). Although prions in the brains of diseased, SSBP/1-infected TgEq were undetectable using RK-OvV, their subsequent passage to TgOvV resulted in titers comparable to those in TgOv or sheep infected with SSBP/1 (Fig. 3B). These results are consistent with transient adaptation of SSBP/1 in TgEq by NAPA to produce Eq-SSBP/1, to which RK-OvV cells are refractory, and concomitant clearance of the SSBP/1 inoculum, whereas the subsequently elevated titers in TgOvV are consistent with efficient amplification of NAPA horse prions in TgEq. In contrast to deer prions produced by adaptation of mouse RML or SSBP/1 in TgD, referred to as D-RML and D-SSBP/1 (9, 10), D-TME prions from diseased, TME-infected TgD produced by NAPA failed to infect RK-D (Fig. 3C). These results are in accordance with replication of deer prions by NAPA in the brains of TME-infected TgD and the interpretation that RK-D cells are susceptible to deer-adapted prions but refractory to nonadapted deer prions.
Fig. 3.
Prion replication during NAPA. Cells were challenged with dilutions of 10% brain homogenates in PBS. Mean cell counts were assessed from three plates; error bars, SEM. (A, Upper) RK-13 cells expressing mouse (RK-M) infected with RML from diseased FVB mice (blue squares) or asymptomatic TgEq (blue filled red circles). (Lower) RK-13 cells expressing deer PrPC (RK-D) infected with deer CWD (blue squares) or asymptomatic, CWD-challenged TgEq (blue filled red circles). (B, Upper) Titration of SSBP/1 from sheep (blue squares) or two SSBP/1-infected TgOvV (dotted blue squares) in RK-13 cells expressing sheep (RK-OvV). (Lower) Titration of two TgOvV brains infected with NAPA horse prions in RK-OvV (red filled black squares) and NAPA horse prions from the brain of a diseased TgEq infected with SSBP/1 (clear black squares). (C) Susceptibility of RK-D cells to CWD, and RML and SSBP/1 prions adapted in TgD (D-RML and D-SSBP/1) and to NAPA deer prions from the brain of a TME-infected diseased TgD. Error bars, SEM of at least five replicates. (D and E) Western blots of TgD, TgM, and mink brain extracts probed with mAbs 6H4 and PRC1 that distinguish prions from the species of origin (blue) and adaptation (red). Samples treated (+) or not (–) with PK are shown beneath each lane. In D, PRC1 reacts with CWD but not TME or NAPA deer prions; in E, PRC1 reacts only with PMCA-Eq-RML.
The availability of species-discriminatory anti-PrP monoclonal antibodies (mAbs) (39) allowed us to differentiate newly replicated prions from input prions using Western blot profiling. TME PrPSc migrated faster than CWD PrPSc, and the migration properties of D-TME prions in TgD and TgMi were related to TME but distinct from CWD (Fig. 3D). mAb PRC1, which recognizes deer and horse but not mink or mouse PrP, failed to recognize PrPSc in TgD or TgMi infected with D-TME due to epitope elimination following treatment with proteinase K (PK), which does not occur in CWD-enciphered PrPSc (Fig. 3D). The reactivity of mAb PRC1 with low PrPSc levels in the brains of asymptomatic TgEq inoculated with PMCA RML-derived horse prions (Fig. 3E) is also in accordance with PrPSc originating from converted EqPrPC.
Discussion
Here we describe three examples of a nonadaptive form of prion replication, which differs from conventional transspecies prion infections where novel prion strains composed of modified PrPSc conformations preferentially replicate in the species of adaptation (9, 10, 20). We also report on the generation and characterization of two new Tg mouse models expressing horse or mink PrP. Our results in TgEq support predictions from structural analyses (27) that horse PrPC is resistant to conformational conversion to PrPSc. Even in the rare event that pathogenic horse prions are produced during infection, replication by NAPA ensures that they are paradoxically not optimized for further conversion of EqPrPC upon passage. The properties of horse PrPC therefore differ significantly from rabbit PrPC, a species incorrectly thought to be resistant to prion infection (15, 36). Nonetheless, several lines of evidence confirm that our description of NAPA in TgEq is not the result of an idiosyncratic resistance of EqPrPC to support prion replication. In addition to NAPA of SSBP/1 and RML by EqPrPC, we describe replication of pathogenic deer prions by NAPA in TgD (Table 1, primary passage of TME prions and serial passage of NAPA deer prions). Here the primary transmission profile of TME prions in TgD differs from that of SSBP/1 in TgEq. It would therefore appear that, like adaptive interspecies prion propagation, NAPA involves a two-step process. Whereas, in both cases, primary transmissions may result in either all inoculated animals developing disease after a prolonged asymptomatic phase, or only a subpopulation of inoculated animals developing disease with variable incubation times, outcomes of secondary transmissions of the resulting prions reflect either adaptive- or nonadaptive amplification. The propagation of TME in TgD by NAPA contrasted the response of TgD to infection with prions from other species including sheep SSBP/1, mouse RML, and cattle BSE, which in all cases resulted in adapted deer prions that caused rapid, synchronous disease upon serial transmission to TgD (9, 10, 40). Our findings are therefore consistent with the notion that NAPA involves particular prion/PrPC interactions during transspecies transmissions. In previous studies, PrPSc conformational changes accompanied emergence of newly adapted prion strains following passage across species barriers (9, 10, 20). NAPA does not entail emergence of new strains or conformational changes in PrPSc but instead produces prions that replicate in the species of prion origination and not the new host. We speculate that NAPA provides an alternate means of prion replication when conditions are unfavorable for fully adaptive propagation. Because horse and deer PrPC are capable of supporting, at least transiently, the replication of several nonpreferred prion conformations, our results contend that transmission barriers are not inevitably determined by selective propagation of novel quasi-species by PrPC in the new host, as postulated by the conformational selection model (21), and that control of prion host range is dictated by selective pressures imposed by PrPSc rather than host-encoded PrPC.
NAPA is also consistent with several previously unexplained transmission phenomena (41). Seminal studies revealed a substantial species barrier of the hamster-adapted scrapie strain 263K on first passage to mice (42). Although Race et al. initially concluded that inoculated 263K prions persisted in the brains of asymptomatic mice (43), their subsequent findings (44, 45), and those of others (33), showed that prions replicated under these conditions. Given the hamster-tropic nature of much of the resulting infectivity, these results appear to represent an early, unrecognized example of NAPA. Despite the generally accepted notion that BSE is the origin of human vCJD, the unusual transmission characteristics of BSE and vCJD have been hard to reconcile in the context of adaptive prion replication (46). Although primary transmission of human-derived BSE prions from vCJD patients to mice expressing human PrP was expected to be efficient, only 7% of Tg35 mice with twofold overexpression of human PrP with methionine (M) at residue 129 (HuPrP-M129) were susceptible to disease, with variable, prolonged incubation times (47). Here, and subsequently (48), the majority of vCJD-infected Tg(HuPrP-M129) mice remained asymptomatic, despite prion replication indicated by PrPSc accumulation, a situation reminiscent of NAPA (Fig. 1C). In subsequently developed tg650 with sixfold HuPrP-M129 expression, vCJD primary transmission rates increased to 67%, possibly as a result of increased transgene expression, with mean incubation times varying between 338 and 628 d and standard errors of the mean ranging from ±15–42 d (49). In a third model, referred to as tg340 with fourfold HuPrP-M129 expression, disease was registered in six of six vCJD-inoculated mice after a protracted ∼630 d mean incubation time with wide variance (50). The lower than expected efficiency of vCJD primary disease transmission is in accordance with the notion that the causative ancestral BSE prions are not optimally adapted for full pathogenic potential in humans. Consistent with suboptimal adaptation, although serial transmission of vCJD prions was accomplished in tg650 and tg340, incubation times remained protracted and variable (49, 50). In contrast, the preferential transmission of human vCJD prions to Tg mice expressing ancestral bovine PrPC [Tg(BoPrP)] without an obvious transmission barrier, producing neuropathological and PrPSc molecular profiles corresponding to BSE, was equally surprising and at odds with adaptive prion transmission (46). These unexpected vCJD transmission properties were subsequently confirmed using independently produced TgBoPrP (50, 51). Confirming the shared properties of BSE/vCJD prions, the transmission profile of vCJD in Tg(HuPrP) and Tg(BoPrP) corresponded to that of BSE prions previously passaged in Tg(HuPrP) mice (51). The overlapping denaturation profiles of PrPSc in the brains of diseased cattle and humans (52, 53) are consistent with shared conformations of vCJD and BSE prions, which is also in accordance with NAPA.
Finally, our findings also constitute new elements that feed the debate surrounding the role of the β2–α2 PrP loop in the development of age-related prion disease. Development of spontaneous disease in Tg mice expressing mouse PrPC containing rigid β2–α2 loops from these species (28, 29) present a conundrum in the face of structural predictions of EqPrPC stability (27) and our findings that expression of these same rigid loops in the context of deer PrP (3) and horse PrP (Table 1) does not result in spontaneous neurodegeneration. Our previous studies in which we correlated the effects of natural, protective polymorphisms in α-helix3 with increased stability of a discontinuous epitope comprised of residues in β2–α2 and α-helix3 (54) confirmed structural predictions that motif plasticity is a crucial factor during PrPC conversion (25, 27). We speculate that although expression of PrP constructs with inherently rigid β2–α2 loops may potentiate spontaneous PrP conversion, this effect is counterbalanced by long-range interactions with α-helix3 resident amino acids that otherwise stabilize and prevent PrPC conversion.
Materials and Methods
Tg Mice.
Animal work was performed in compliance with the requirements of University Institutional Animal Care and Use Committees. Tg mice were developed and characterized as previously described (3) using the MoPrP.Xho expression vector.
Inocula.
SSBP/1 was from infected Cheviot sheep; Stetsonville TME was prepared as described (31). RML mouse prions were passaged in wild-type FVB mice. BSE and BASE field cases were supplied by the Laboratorio Central de Veterinaria. Atypical scrapie was a field case diagnosed at the Centre for Research into Animal Health (CReSA). CWD isolates were from naturally affected deer or elk. Ten percent brain homogenates were prepared, and mice were inoculated and diagnosed as described (10).
Cell Lines and Prion Titrations.
RK13 cells (cat. no. CCL-37; ATCC) were engineered to stably express elk, deer, mouse, or ovine PrP using pIRESpuro3 (Clontech). Cells highly sensitive to infection were isolated by cloning. Titers of CWD, RML, and SSBP/1 prions were assessed in RKD, RKM, and RKOv-V cell lines, respectively, as described (55).
Western blotting of 10% brain homogenates normalized for protein content were performed as previously described (30).
PMCA.
SSBP/1, RML, and BSE seeded horse prions (Eq-SSBP/1, Eq-RML, and Eq-BSE) were generated using horse brain homogenate in sPMCA as described previously (10).
Conformational Stability Assay.
Brain homogenates containing 10 μg total protein were incubated with various concentrations of GdnHCl for 1 h at room temperature, followed by PK digestion, and analyzed as described (55).
Acknowledgments
We thank members of the Prion Research Center (PRC) for critical discussions and feedback, including Dr. Julie Moreno, and in particular the greatly missed Dr. Richard Bessen; Tanya Seward for technical assistance during the early stages of this project; and our colleagues at Prionics for mAb 6H4. This work was supported by National Institutes of Health Grants R01 NS040334 and P01 AI077774.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. I.V.B. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611891114/-/DCSupplemental.
References
- 1.Prusiner SB. Prions (Les Prix Nobel Lecture) In: Frängsmyr T, editor. Les Prix Nobel. Almqvist & Wiksell International; Stockholm, Sweden: 1998. pp. 268–323. [Google Scholar]
- 2.Baskakov IV. The many shades of prion strain adaptation. Prion. 2014;8(2):27836. doi: 10.4161/pri.27836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Browning SR, et al. Transmission of prions from mule deer and elk with chronic wasting disease to transgenic mice expressing cervid PrP. J Virol. 2004;78(23):13345–13350. doi: 10.1128/JVI.78.23.13345-13350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibbs CJ, Jr, Gajdusek DC. Experimental subacute spongiform virus encephalopathies in primates and other laboratory animals. Science. 1973;182(4107):67–68. doi: 10.1126/science.182.4107.67. [DOI] [PubMed] [Google Scholar]
- 5.Gibbs CJ, Jr, Gajdusek DC, Amyx H. Strain variation in the viruses of Creutzfeldt-Jakob disease and kuru. In: Prusiner SB, Hadlow WJ, editors. Slow Transmissible Diseases of the Nervous System. Vol 2. Academic; New York: 1979. pp. 87–110. [Google Scholar]
- 6.Di Bari MA, et al. The bank vole (Myodes glareolus) as a sensitive bioassay for sheep scrapie. J Gen Virol. 2008;89(Pt 12):2975–2985. doi: 10.1099/vir.0.2008/005520-0. [DOI] [PubMed] [Google Scholar]
- 7.Nicot S, Baron T. Strain-specific barriers against bovine prions in hamsters. J Virol. 2011;85(4):1906–1908. doi: 10.1128/JVI.01872-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruce ME, et al. Strain characterization of natural sheep scrapie and comparison with BSE. J Gen Virol. 2002;83(Pt 3):695–704. doi: 10.1099/0022-1317-83-3-695. [DOI] [PubMed] [Google Scholar]
- 9.Green KM, et al. The elk PRNP codon 132 polymorphism controls cervid and scrapie prion propagation. J Gen Virol. 2008;89(Pt 2):598–608. doi: 10.1099/vir.0.83168-0. [DOI] [PubMed] [Google Scholar]
- 10.Green KM, et al. Accelerated high fidelity prion amplification within and across prion species barriers. PLoS Pathog. 2008;4(8):e1000139. doi: 10.1371/journal.ppat.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimberlin RH, Cole S, Walker CA. Temporary and permanent modifications to a single strain of mouse scrapie on transmission to rats and hamsters. J Gen Virol. 1987;68(Pt 7):1875–1881. doi: 10.1099/0022-1317-68-7-1875. [DOI] [PubMed] [Google Scholar]
- 12.Scott M, et al. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59(5):847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 13.Lee YH, et al. Experimental chronic wasting disease in wild type VM mice. J Vet Med Sci. 2013;75(8):1107–1110. doi: 10.1292/jvms.13-0018. [DOI] [PubMed] [Google Scholar]
- 14.Telling GC. Transgenic mouse models and prion strains. Top Curr Chem. 2011;305:79–99. doi: 10.1007/128_2011_166. [DOI] [PubMed] [Google Scholar]
- 15.Vidal E, et al. Transgenic mouse bioassay: Evidence that rabbits are susceptible to a variety of prion isolates. PLoS Pathog. 2015;11(8):e1004977. doi: 10.1371/journal.ppat.1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espinosa JC, et al. Transgenic mice expressing porcine prion protein resistant to classical scrapie but susceptible to sheep bovine spongiform encephalopathy and atypical scrapie. Emerg Infect Dis. 2009;15(8):1214–1221. doi: 10.3201/eid1508.081218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Telling GC, et al. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274(5295):2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 18.Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol. 1994;68(12):7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dickinson AG. Scrapie in sheep and goats. In: Kimberlin RH, editor. Slow Virus Diseases of Animals and Man. North-Holland Publishing; Amsterdam: 1976. pp. 209–241. [Google Scholar]
- 20.Peretz D, et al. A change in the conformation of prions accompanies the emergence of a new prion strain. Neuron. 2002;34(6):921–932. doi: 10.1016/s0896-6273(02)00726-2. [DOI] [PubMed] [Google Scholar]
- 21.Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318(5852):930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 22.Sigurdson CJ, et al. A molecular switch controls interspecies prion disease transmission in mice. J Clin Invest. 2010;120(7):2590–2599. doi: 10.1172/JCI42051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurt TD, et al. Human prion protein sequence elements impede cross-species chronic wasting disease transmission. J Clin Invest. 2015;125(4):1485–1496. doi: 10.1172/JCI79408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gossert AD, Bonjour S, Lysek DA, Fiorito F, Wüthrich K. Prion protein NMR structures of elk and of mouse/elk hybrids. Proc Natl Acad Sci USA. 2005;102(3):646–650. doi: 10.1073/pnas.0409008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christen B, Hornemann S, Damberger FF, Wüthrich K. Prion protein NMR structure from tammar wallaby (Macropus eugenii) shows that the beta2-alpha2 loop is modulated by long-range sequence effects. J Mol Biol. 2009;389(5):833–845. doi: 10.1016/j.jmb.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 26.Khan MQ, et al. Prion disease susceptibility is affected by beta-structure folding propensity and local side-chain interactions in PrP. Proc Natl Acad Sci USA. 2010;107(46):19808–19813. doi: 10.1073/pnas.1005267107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez DR, Damberger FF, Wüthrich K. Horse prion protein NMR structure and comparisons with related variants of the mouse prion protein. J Mol Biol. 2010;400(2):121–128. doi: 10.1016/j.jmb.2010.04.066. [DOI] [PubMed] [Google Scholar]
- 28.Sigurdson CJ, et al. De novo generation of a transmissible spongiform encephalopathy by mouse transgenesis. Proc Natl Acad Sci USA. 2009;106(1):304–309. doi: 10.1073/pnas.0810680105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sigurdson CJ, et al. Spongiform encephalopathy in transgenic mice expressing a point mutation in the β2-α2 loop of the prion protein. J Neurosci. 2011;31(39):13840–13847. doi: 10.1523/JNEUROSCI.3504-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saijo E, et al. Epigenetic dominance of prion conformers. PLoS Pathog. 2013;9(10):e1003692. doi: 10.1371/journal.ppat.1003692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartz JC, Bessen RA, McKenzie D, Marsh RF, Aiken JM. Adaptation and selection of prion protein strain conformations following interspecies transmission of transmissible mink encephalopathy. J Virol. 2000;74(12):5542–5547. doi: 10.1128/jvi.74.12.5542-5547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Windl O, et al. Breaking an absolute species barrier: Transgenic mice expressing the mink PrP gene are susceptible to transmissible mink encephalopathy. J Virol. 2005;79(23):14971–14975. doi: 10.1128/JVI.79.23.14971-14975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill AF, et al. Species-barrier-independent prion replication in apparently resistant species. Proc Natl Acad Sci USA. 2000;97(18):10248–10253. doi: 10.1073/pnas.97.18.10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411(6839):810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 35.Castilla J, et al. Crossing the species barrier by PrP(Sc) replication in vitro generates unique infectious prions. Cell. 2008;134(5):757–768. doi: 10.1016/j.cell.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chianini F, et al. Rabbits are not resistant to prion infection. Proc Natl Acad Sci USA. 2012;109(13):5080–5085. doi: 10.1073/pnas.1120076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bian J, et al. Cell-based quantification of chronic wasting disease prions. J Virol. 2010;84(16):8322–8326. doi: 10.1128/JVI.00633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vilette D, et al. Ex vivo propagation of infectious sheep scrapie agent in heterologous epithelial cells expressing ovine prion protein. Proc Natl Acad Sci USA. 2001;98(7):4055–4059. doi: 10.1073/pnas.061337998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang HE, et al. Characterization of conformation-dependent prion protein epitopes. J Biol Chem. 2012;287(44):37219–37232. doi: 10.1074/jbc.M112.395921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vickery CM, et al. Assessing the susceptibility of transgenic mice overexpressing deer prion protein to bovine spongiform encephalopathy. J Virol. 2014;88(3):1830–1833. doi: 10.1128/JVI.02762-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nonno R, et al. Efficient transmission and characterization of Creutzfeldt-Jakob disease strains in bank voles. PLoS Pathog. 2006;2(2):e12. doi: 10.1371/journal.ppat.0020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimberlin RH, Walker CA, Fraser H. The genomic identity of different strains of mouse scrapie is expressed in hamsters and preserved on reisolation in mice. J Gen Virol. 1989;70(Pt 8):2017–2025. doi: 10.1099/0022-1317-70-8-2017. [DOI] [PubMed] [Google Scholar]
- 43.Race R, Chesebro B. Scrapie infectivity found in resistant species. Nature. 1998;392(6678):770. doi: 10.1038/33834. [DOI] [PubMed] [Google Scholar]
- 44.Race R, Raines A, Raymond GJ, Caughey B, Chesebro B. Long-term subclinical carrier state precedes scrapie replication and adaptation in a resistant species: Analogies to bovine spongiform encephalopathy and variant Creutzfeldt-Jakob disease in humans. J Virol. 2001;75(21):10106–10112. doi: 10.1128/JVI.75.21.10106-10112.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Race R, et al. Subclinical scrapie infection in a resistant species: Persistence, replication, and adaptation of infectivity during four passages. J Infect Dis. 2002;186(Suppl 2):S166–S170. doi: 10.1086/344267. [DOI] [PubMed] [Google Scholar]
- 46.Scott MR, et al. Compelling transgenetic evidence for transmission of bovine spongiform encephalopathy prions to humans. Proc Natl Acad Sci USA. 1999;96(26):15137–15142. doi: 10.1073/pnas.96.26.15137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asante EA, et al. BSE prions propagate as either variant CJD-like or sporadic CJD-like prion strains in transgenic mice expressing human prion protein. EMBO J. 2002;21(23):6358–6366. doi: 10.1093/emboj/cdf653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wadsworth JD, et al. Human prion protein with valine 129 prevents expression of variant CJD phenotype. Science. 2004;306(5702):1793–1796. doi: 10.1126/science.1103932. [DOI] [PubMed] [Google Scholar]
- 49.Béringue V, et al. Prominent and persistent extraneural infection in human PrP transgenic mice infected with variant CJD. PLoS One. 2008;3(1):e1419. doi: 10.1371/journal.pone.0001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Padilla D, et al. Sheep and goat BSE propagate more efficiently than cattle BSE in human PrP transgenic mice. PLoS Pathog. 2011;7(3):e1001319. doi: 10.1371/journal.ppat.1001319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torres JM, et al. Elements modulating the prion species barrier and its passage consequences. PLoS One. 2014;9(3):e89722. doi: 10.1371/journal.pone.0089722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watts JC, et al. Evidence that bank vole PrP is a universal acceptor for prions. PLoS Pathog. 2014;10(4):e1003990. doi: 10.1371/journal.ppat.1003990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Safar JG, et al. Measuring prions causing bovine spongiform encephalopathy or chronic wasting disease by immunoassays and transgenic mice. Nat Biotechnol. 2002;20(11):1147–1150. doi: 10.1038/nbt748. [DOI] [PubMed] [Google Scholar]
- 54.Angers R, et al. Structural effects of PrP polymorphisms on intra- and interspecies prion transmission. Proc Natl Acad Sci USA. 2014;111(30):11169–11174. doi: 10.1073/pnas.1404739111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bian J, Kang HE, Telling GC. Quinacrine promotes replication and conformational mutation of chronic wasting disease prions. Proc Natl Acad Sci USA. 2014;111(16):6028–6033. doi: 10.1073/pnas.1322377111. [DOI] [PMC free article] [PubMed] [Google Scholar]