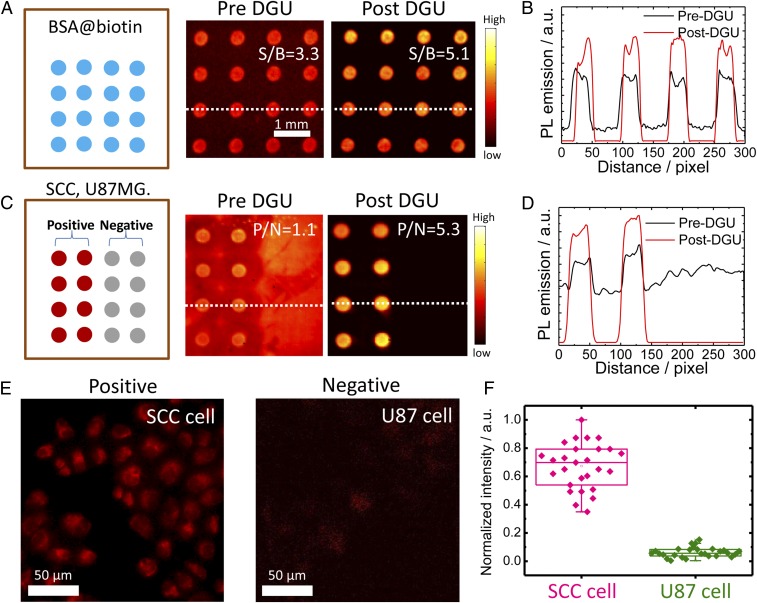
Fig. 2.
The microarray assay screening method developed to test the conjugates. (A) Assay analysis of DGU fraction of SA@IR-FGP. BSA-biotin was printed on the plasmonic fluorescence-enhancing gold slide, after which the pre/post-DGU conjugates were incubated on the slide. Finally, the slide was scanned by a 10× magnification NIR-II setup with 808-nm excitation and a 1,100-nm long-pass emission filter. (B) Cross-sectional intensity profile of the spot signal. (C and D) Assay analysis of the DGU fractions of Erb@IR-FGP. SCC and U87 cell lysates were printed on plasmonic fluorescence-enhancing gold slides for testing the conjugate Erb@IR-FGP. The slide was finally scanned with a 10× magnification NIR-II setup with 808-nm excitation and a 1,100-nm long-pass emission filter. Note that the gold slide has a certain autofluorescence under the testing condition. (E) Targeted cell staining by Erb@IR-FGP (SCC and U87 cell lines as positive and negative controls) scanned by a home-built microscope with 785-nm excitation and a 1,050-nm long-pass emission filter. (F) PL intensity statistic of SCC and U87 cells.