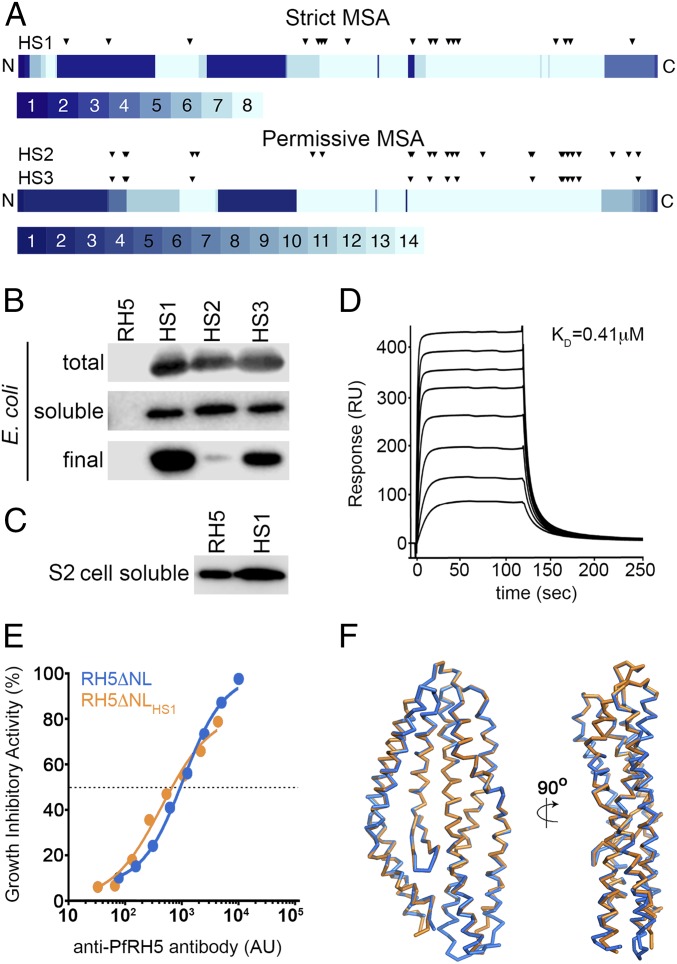
Fig. 1.
Design of an E. coli-expressible PfRH5 variant. (A) Eighty percent of mutations in all three designed variants are located in the C-terminal half of PfRH5, where most of the aligned sequences contribute information on sequence diversity. The PfRH5 sequence is schematically shown from N to C terminus, and each position is colored according to the number of unique sequences contributing to the strict and permissive alignments, ranging from 1 to 8 and from 1 to 14, respectively. The locations of the mutations in each designed variant relative to wild-type PfRH5ΔNL are indicated by triangles. (B) Expression levels of PfRH5ΔNL (RH5), PfRH5ΔNLHS1 (HS1), PfRH5ΔNLHS2 (HS2), and PfRH5ΔNLHS3 (HS3) from E. coli. “Total” is whole lysed cells, “soluble” is material after cell lysis and clarification, and “final” is after immobilized metal ion affinity chromatography and size exclusion chromatography. (C) Expression levels of PfRH5ΔNL (RH5) and PfRH5ΔNLHS1 (HS1) secreted in the cell supernatants from a stable Drosophila melanogaster Schneider 2 (S2) cell line. (D) Surface plasmon resonance analysis of the binding of PfRH5ΔNLHS1 to basigin, with twofold dilutions of PfRH5ΔNLHS1 from a maximal concentration of 8 μM. (E) In vitro GIA of purified IgG against 3D7 clone parasites from mice immunized with either PfRH5ΔNL or PfRH5ΔNLHS1. The anti-PfRH5 antibody response was measured for each sample of purified IgG and is plotted against the measured level of GIA. The dashed line indicates 50% GIA, and each GIA data point represents the mean of each sample tested in triplicate. (F) The structure of PfRH5ΔNLHS1:9AD4 (orange) overlaid on the structure of PfRH5ΔNL:9AD4 (blue).