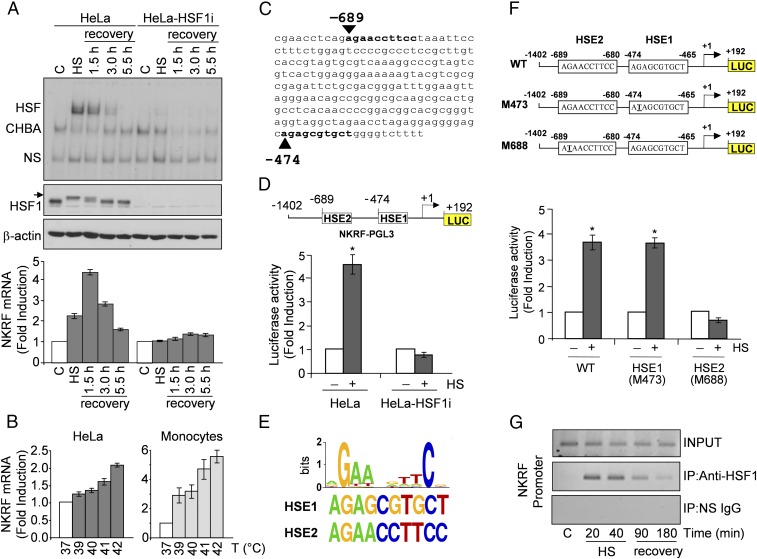
Fig. 1.
HSF1 directly regulates NKRF expression during temperature increase. (A) HSF1 DNA-binding activity (gel-shift analysis, Top) and phosphorylation (immunoblot, IB, Middle) and NKRF-mRNA levels (qPCR, Bottom) of whole-cell protein or RNA extracts from HeLa and HeLa-HSF1i cells exposed to heat stress (HS, 43 °C, 40 min) and allowed to recover at 37 °C for the indicated times. CHBA, constitutive HSE-binding activity; HSF, HSF/DNA complexes; NS, nonspecific protein–DNA interactions. Arrow indicates hyperphosphorylated HSF1. (B) NKRF mRNA levels in HeLa cells or human peripheral-blood monocytes incubated at febrile temperatures (2 h). (C) NKRF promoter putative HSF1-binding sites (bold) identified by TFSearch. (D) NKRF promoter reporter analysis in HeLa and HeLa-HSF1i cells transfected with NKRF–PGL3 (Top) and heat stressed (6 h recovery). Putative HSF1-binding sites (HSE1, HSE2) identified in C are shown; numbers indicate positions relative to TSS (+1). (E, Top) WebLogo-generated HSF1 consensus motif. (Bottom) NKRF promoter HSE1 and HSE2 sequences. (F) Wild-type (WT) and point-mutated (G-to-T) (M473, M688) constructs (Top) were used for reporter analysis in HeLa cells treated as in D (Bottom). (G) ChIP analysis of HSF1 binding to the NKRF promoter in HeLa cells treated as in A. ChIP-enriched DNAs using preimmune (IP:NS-IgG) or anti-HSF1 (IP:anti-HSF1) serum and input DNAs are shown. Error bars indicate ±SD. *P < 0.05.