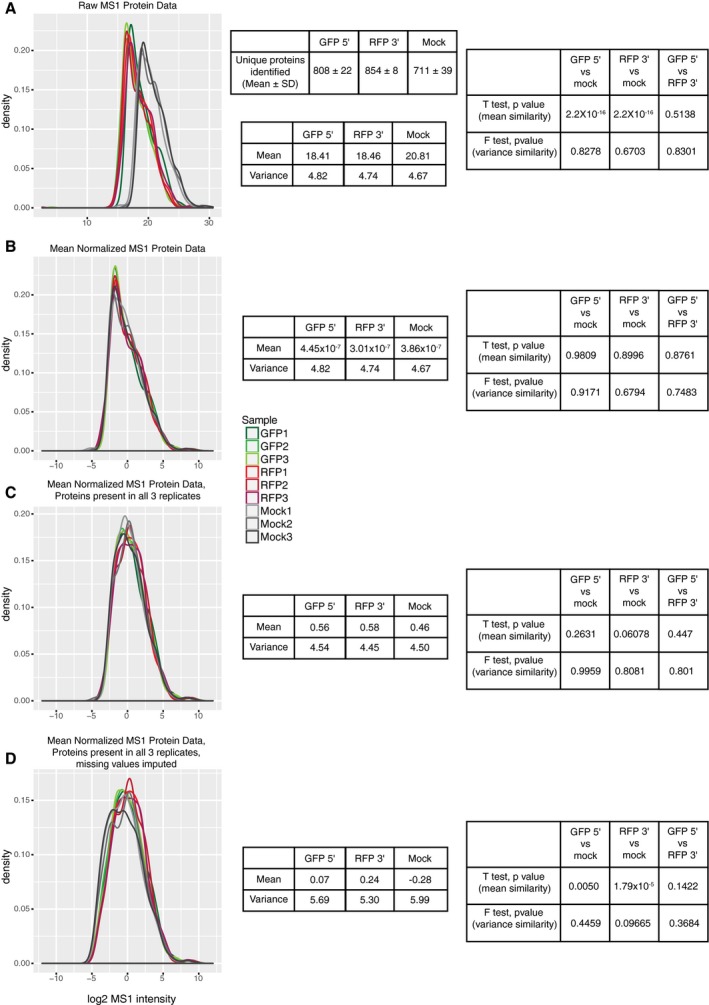
The distribution of summed MS1 intensities from all peptides corresponding to each protein identified in each mass spectrometry run are plotted. The distribution of MS1 intensities is plotted for all nine samples (triplicate 5′ IPs, triplicate 3′ IPs, and triplicate mock IPs). Also shown is a table displaying the mean and standard deviation for the number of unique proteins identified from the triplicate IPs. The distributions are plotted for (A) raw MS1 intensities, (B) mean normalized (see
Materials and Methods) intensities, (C) mean normalized intensities filtered for proteins present in all three samples from at least one condition, and (D) mean normalized intensities filtered for proteins present in all three samples from at least one condition with missing values (i.e. proteins present in one condition but not the other) imputed (see
Materials and Methods). The mean and variance are shown in the table next to the distributions represent the mean and variance from averaging the triplicate datasets.
t‐tests to test for differences in the mean and
F‐tests to test for differences in the variance for each dataset are also shown. These data demonstrate that both raw and normalized datasets do not vary in their variances as
F‐tests between the datasets are not significant. The 5′ and 3′ IPs show highly similar mean MS1 intensity values and distributions in the raw data. The mock IPs display a mean shifted toward larger MS1 intensities. Thus, to allow for comparison between 5′, 3′ IPs, and mock IPs, the data were mean normalized (as described in the
Materials and Methods) to center the distributions of the datasets around zero, allowing the samples to be compared to one another. As the variance was not significantly different between any of the datasets, no adjustment of the variance was made. After mean normalization, the MS1 intensity distributions and variances are all similar, allowing direct comparison of protein enrichment within samples as well as
t‐tests between each protein in each sample. Note that dataset mean and variance values do not differ between samples until imputation of missing values to allow for statistical analysis between samples.