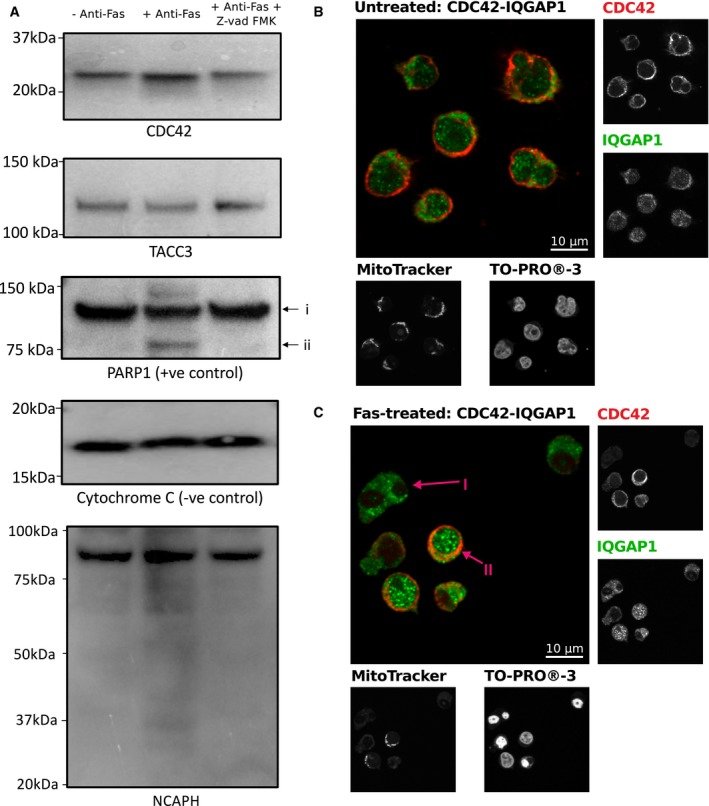
Figure 6. Western blot‐based and co‐localization investigation of protein‐specific proteolysis of known caspase targets which undergo changes in the interactome.
-
AWestern blotting analysis of TACC3, CDC42 and NCAPH with CYC1 (cytochrome c1) and PARP1 as controls. Lanes correspond to: (1) with no treatment, (2) in response to 4‐h Fas treatment, and (3) in response to 4‐h Fas stimulation with prior treatment with Z‐vad‐FMK. In response to treatment, TACC3, CDC42 and NCAPH do not undergo detectible cleavage. Examination of the low mass region of NCAPH supports some, albeit very minor, detectible proteolysis in a caspase‐dependent manner after 4‐h Fas stimulation.
-
B, CCo‐localization analysis of CDC42 and its interaction partner IQGAP1 without (B) and with 4‐h Fas stimulation (C). Anti‐CDC42 and anti‐IQGAP1 staining shows co‐localization of CDC42 and IQGAP1 within cells which have maintained mitochondrial membrane potential, as determined by the retention of MitoTracker, and non‐condensed nuclei as determined by TO‐PRO‐3 iodide. Arrow (I) highlights an example of apoptosis‐committed cell showing loss of CDC42, loss of mitochondrial membrane potential, and condense nuclei. Arrow (II) highlights an example of a pre‐apoptotic cell which has maintained CDC42, mitochondrial membrane potential, and nuclei is non‐condensed.
