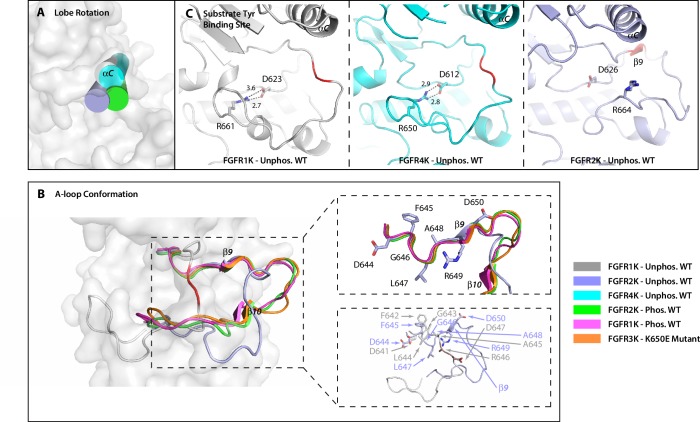
Figure 2. The crystal structure of unphosphorylated WT FGFR2K is in a partially active conformation.
(A) The orientation of the αC helix (rendered as a cylinder) in the crystal structure of unphosphorylated WT FGFR2K is similar to those seen in crystal structures of the A-loop phosphorylated FGFR1K and FGFR2K. (B) The conformation of first seven residues of the A-loop (DFGLARD) of the unphosphorylated WT FGFR2K resembles those of phosphorylated activated FGFR1K and FGFR2K. Note that as in these latter phosphorylated activated FGFRK structures, the unphosphorylated FGFR2K features the β9 strand which pairs with the catalytic loop. (C) The substrate tyrosine binding site in the unphosphorylated FGFR2K is fully accessible which is in stark contrast to that in the unphosphorylated FGFR1K and FGFR4K in which a prominent salt-bridge between an FGFR-invariant arginine (R661 in FGFR1K, R664 in FGFR2K and R655 in FGFR4K) and the catalytic base (D623 in FGFR1K, D626 in FGFR2K and D617 in FGFR4K) blocks the active site. Hydrogen bonds are shown as dashed lines with the distances given in Å. The A-loop section in the unphosphorylated FGFR1K and FGFR2K that forms a β9 strand upon A-loop phosphorylation is highlighted in red. Note that the β9 strand is already present in the unphosphorylated FGFR2K implying that this structure is in a partially active state.