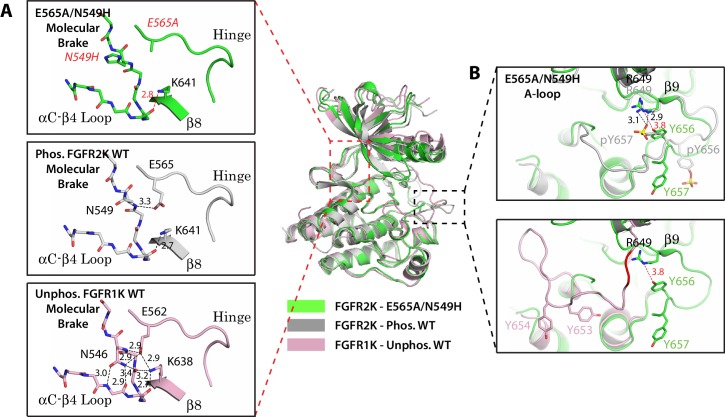
Figure 3. Two mutations within the molecular brake disrupt the hydrogen bonding at the molecular brake and drive the A-loop into the active conformation.
Crystal structures are displayed for the E565A/N549H double mutant of FGFR2K (in green), phosphorylated activated WT FGFR2K (PDB ID: 2PVF [Chen et al., 2007], in grey), and unphosphorylated autoinhibited WT FGFR1K (PDB ID: 1FGK [Mohammadi et al., 1996], in pink). (A) Close-up views of the molecular brake region for each of the three structures. Note that reminiscent of the A-loop phosphorylated activated FGFR2K, the molecular brake is disengaged in the unphosphorylated E565A/N549K structure. The two mutated residues, namely N549H and E565A, are labeled with red letters. (B) Comparison of the A-loop conformations for the three structures. The top panel shows a zoomed-in view of the A-loop conformation for E565A/N549K superimposed onto that of phosphorylated FGFR2K. The bottom panel shows a zoomed-in view of the A-loop conformation for E565A/N549K superimposed onto that of unphosphorylated WT FGFR1K. Backbone residues in unphosphorylated FGFR1K within the A-loop that form the β9 strand after A-loop phosphorylation are highlighted in red. Hydrogen bonds are shown as dashed black (WT structures) and red (mutant structure) lines with distances displayed in Å. Side chains of relevant residues are shown as sticks. Atom colorings are the same as in Figure 1.