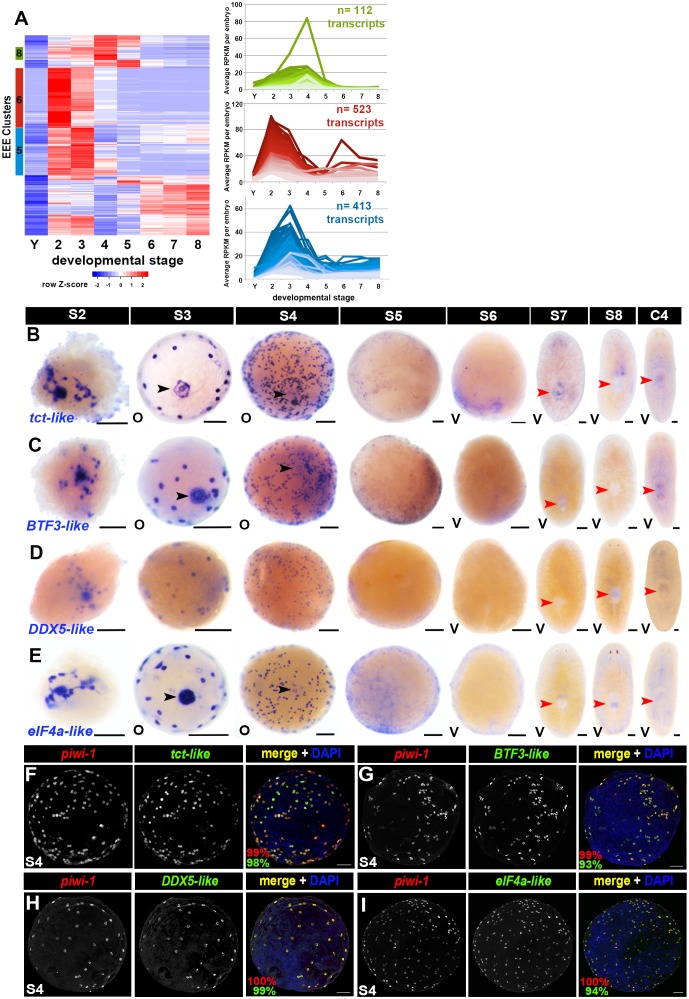
Figure 5. Early-embryo-enriched transcripts are downregulated as organogenesis begins.
(A) Hierarchical clustering of S2–S4-enriched transcripts (n = 1,756) using scaled RPKM data. Left: Heat map. Y, yolk. Colored bars (left) denote Clusters 5, 6 and 8 containing early-embryo-enriched (EEE) transcripts. Cluster f5 sequences (blue, n = 413) were expressed at roughly equivalent levels during S2 and S3, with 66% (n = 275) transcripts showing five-fold or greater declines in average expression values between S3 and S5. Cluster 6 sequences (red, n = 523) exhibited maximal expression during S2, and average expression levels declined more than five-fold between S2 and S4 for 81% (n = 426) of these transcripts. Cluster 8 sequences (green, n = 112) showed peak expression during S4, with 52% (n = 60) of the transcripts showing five-fold or greater declines in average expression values by S5. Right: Normalized expression trends for EEE transcripts in Clusters 5 (blue), 6 (red) and 8 (green) plotted as a function of developmental time. Median 50% of transcripts based on expression maxima are plotted. (B–E) Colorimetric WISH depicting expression of the EEE transcripts tct-like (B), BTF3-like (C), DDX5-like (D) and eIF4a-like (E) (blue) in S2–S8 embryos and C4 asexual adults. Black arrowheads: temporary embryonic pharynx. Red arrowheads: definitive pharynx. O, oral; V, ventral. Scale bars: 100 µm. (F–I) EEE transcripts were expressed throughout the piwi-1+ compartment in S3–S4 embryos. Fluorescent double WISH with riboprobes against piwi-1 (red) and the EEE transcripts tct-like (F), BTF3-like (G), DDX5-like (H) and eIF4a-like (I) (green) in S4 embryos. Percentage piwi-1+ cells coexpressing the indicated EEE marker (red) and percentage EEE+ cells coexpressing piwi-1 (green) appear in the lower left corner of merged images. (F) n = 895 piwi-1+ cells, n = 905 tct-like+ cells, n = 7 S3–S4 embryos. (G) n = 692 piwi-1+ cells, n = 728 BTF3+ cells, n = 6 S3–S4 embryos. (H) n = 676 piwi-1+ cells, n = 681 DDX5-like+ cells, n = 5 S3–S4 embryos. (I) n = 312 piwi1+ cells, n = 332 eIF4a+ cells, n = 4 S3–S4 embryos.
DOI: http://dx.doi.org/10.7554/eLife.21052.047