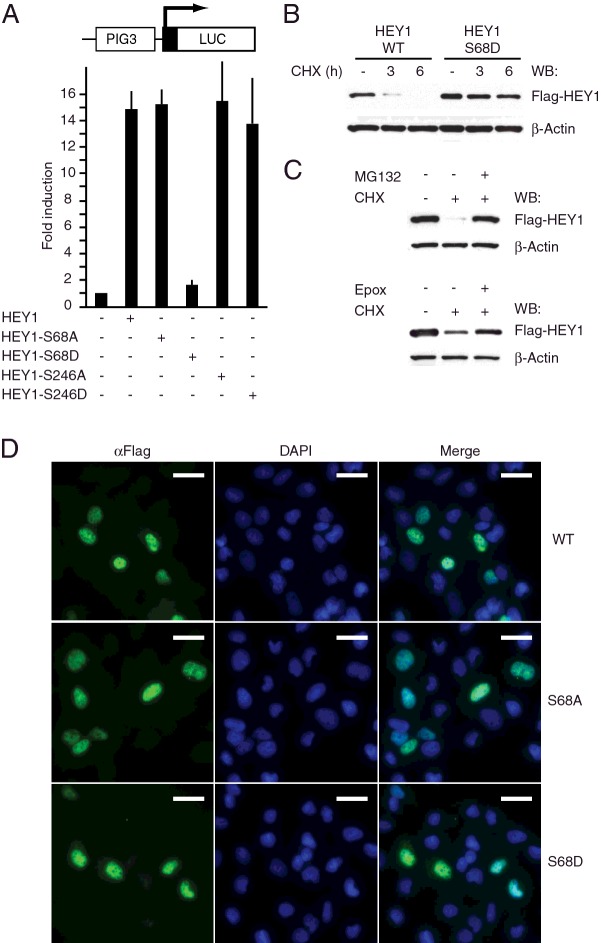
Figure 2. Simulation of HEY1 phosphorylation at residue Ser-68 inhibits its ability to enhance p53 transcriptional activity.
(A) U2OS cells were transfected with 100 ng of PIG3-LUC and 200 ng of expression vectors for HEY1, HEY1-S68A, HEY1-S68D, HEY1-S246A or HEY1-S246D. After transfection, cells were incubated 24 h. Subsequently cell lysates were assayed using a dual luciferase reporter system. Normalized values are expressed relative to the activity of the reporter in the absence of HEY1. The results shown represent the averages of results of three independent experiments assayed in duplicate + S.D. (B) Simulation of HEY1 phosphorylation at residue S68 increases protein stability. U2OS cells were transfected with expression vectors for Flag-tagged HEY1 or HEY1-S68D. Twenty-four hours after transfection cells were treated with cycloheximide (CHX, 10 μg/ml) and HEY1 protein levels were analysed by western blotting at 0, 3 and 6 h after CHX addition. (C) HEY1 is degraded via proteasome. Degradation of HEY1 protein following cycloheximide treatment was prevented by addition of different proteasome inhibitors; MG132 (25 μM) and Epoxomicin (Epox, 1 μM). Anti-β-actin antibody was used as a loading control for all western blots. (D) Simulation of HEY1 phosphorylation does not affect HEY1 nuclear localization. U2OS cells were transfected with expression vectors for Flag-tagged HEY1, HEY1-S68A or HEY-S68D and assayed by indirect immunofluorescence with anti-Flag antibody. The first column shows the indirect immunofluorescence with anti-Flag antibody, the second column shows DAPI staining of DNA and the third column shows the merge image indicating the degree of colocalization. Bars, 20 μm.