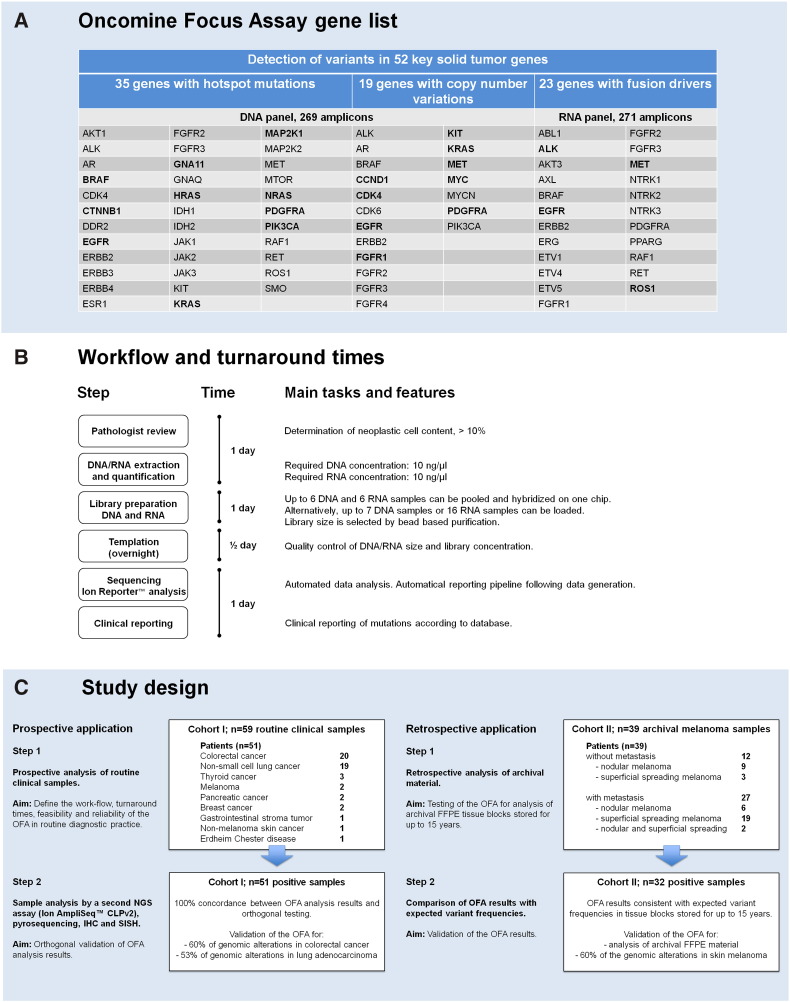
Figure 1.
Molecular profiling with the OFA, a targeted multibiomarker NGS assay. (A) The OFA is a targeted NGS panel that includes 52 solid tumor genes associated with current oncology drugs and published evidence. It enables sequencing of 35 hotspot genes, 19 genes associated with copy number gain, and 23 fusion genes, all in a single workflow using the Ion PGM system. Genes printed in bold were detected in cases of the two cohorts studied here. (B) Workflow and turnaround time for molecular profiling of clinical FFPE samples using targeted NGS with the OFA. (C) Design of this study. The OFA was tested and validated on two different cohorts. Cohort I (left) consisted of 59 routine FFPE samples from 51 patients with various solid tumors that were routinely submitted to the molecular pathology service. Half of these samples were randomly selected for orthogonal validation by alternative tests. In addition, validation was performed by comparing the OFA results with expected mutation frequencies from the literature. Cohort II (right) was an archival cohort of 39 FFPE melanoma samples from 39 patients. Mutation frequencies were retrieved from the literature, and the samples of cohort II were analyzed by the OFA retrospectively. The OFA results of the melanomas in cohort II were consistent with the data that can be expected from large published cohorts of cutaneous melanomas.