FIGURE 5.
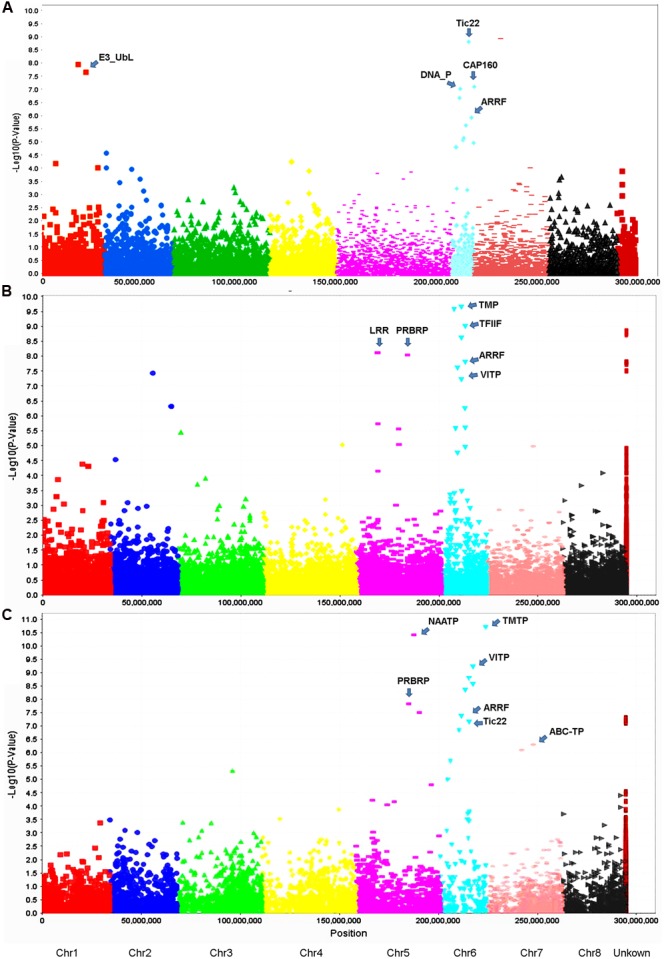
Manhattan and quantile–quantile plots resulting from GWAS using the FreeBayes (A), TASSEL-GBS (B) and UNEAK (C) pipelines in the S & W Seeds population. A false discovery rate of 0.05 was used for significant cutoff (dot lines). Significant markers (above the cutoff lines) associated with VW resistance are listed in Tables 3–5, respectively. The chromosome positions were physical positions (not genetic positions) based on the alignment of alfalfa sequence tags to the reference genome of M. truncatula sequence (Version 3.5). “U” represents unknown chromosome. The abbreviations are putative candidate genes linked to the respective significant marker loci and the arrows indicate their genetic positions in the reference genome. ABC-TP, ABC transporter; ARRF, ankyrin repeat RF-like protein; CAP160, CAP160 repeat protein; DNA_P, DNA primase; E3_UbL, E3 ubiquitin protein ligase XBOS34; LRR, LRR receptor-like kinase family protein; NAATP, neutral amino acid transporter; PRBRP, pumilio-family RNA-binding protein; TFIIF, transcription initiation factor IIF beta subunit; TMP, transmembrane protein; TMTP, transmembrane amino acid transporter family protein; Tic22, Tic22 family protein; VITP, vacuolar iron transporter-like protein.
