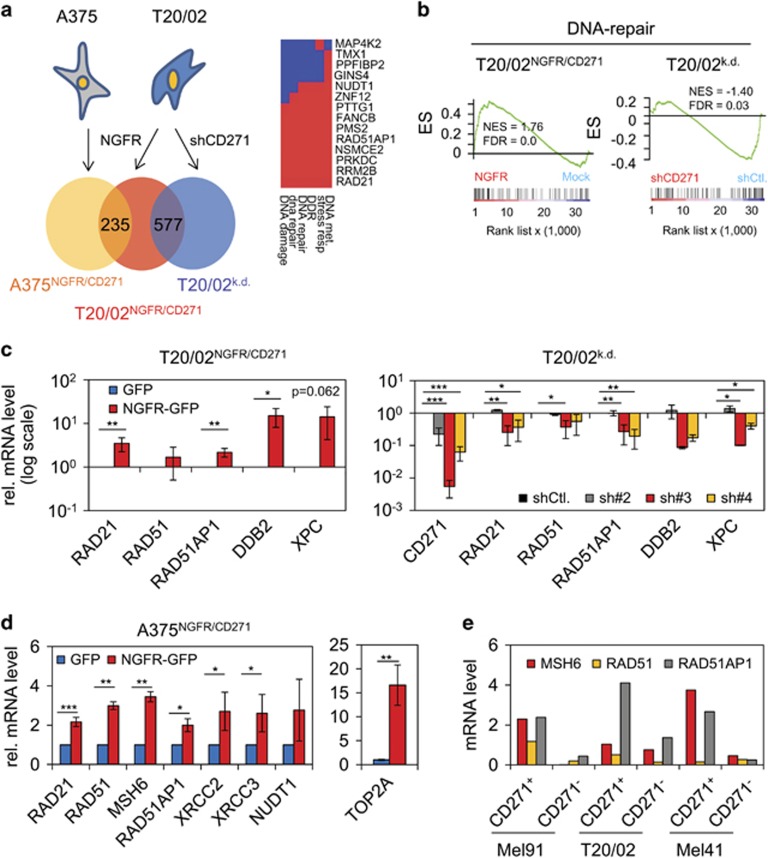
Figure 2.
Integrative analysis of melanoma cells with overexpression and/or knock-down of CD271 links CD271 with DNA repair. (a) Venn diagram depicting commonly upregulated genes induced in A375 and T20/02 cells engineered to stably express CD271 (A375NGFR/CD271, T20/02NGFR/CD271) or stably transfected with a CD271-targeting shRNA (shCD271, T20/02k.d.). Numbers of identified genes found commonly upregulated in A375NGFR/CD271 and T20/02NGFR/CD271 (235 genes) or differentially regulated in T20/02NGFR/CD271 and T20/02k.d. cells (577 genes, comprising up and downregulated genes) are shown. Right panel: DNA repair cluster from DAVID analysis of a set of 340 CD271-responsive genes upregulated in T20/02 cells, red or blue indicates positive or negative GO-contribution, respectively. (b) GSEA of profiling data of T20/02NGFR/CD271 and T20/02k.d. cells using a DNA repair signature (230 genes) as provided by MSigDB.18 (c) Verification of the CD271-dependent regulation of DNA repair genes RAD21, RAD51, RAD51AP1, DNA damage-binding protein 2 (DDB2) and xeroderma pigmentosum genes (XPC) in T20/02NGFR/CD271 (left panel) or cells stably transfected with CD271 specific shRNAs (#2, #3, #4) by qPCR (right panel). Expression levels are shown in logarithmic scale, indicating relative expression levels±s.d. of independent triplicates, compared with cells either expressing GFP or knock-down control (shCtl.). *P⩽0.05; **P⩽0.01; ***P⩽0.001. (d) CD271-induced expression of DNA repair genes in A375NGFR/CD271 cells as determined by qPCR. (e) Expression levels of CD271, RAD51AP1, RAD51 and MSH6 in cells of patient-tumor derived cell strains (Mel91, T20/02 and Mel41) sorted for presence or absence of CD271 by fluorescence-assisted cell sorting (FACS). Shown are three independent experiments.