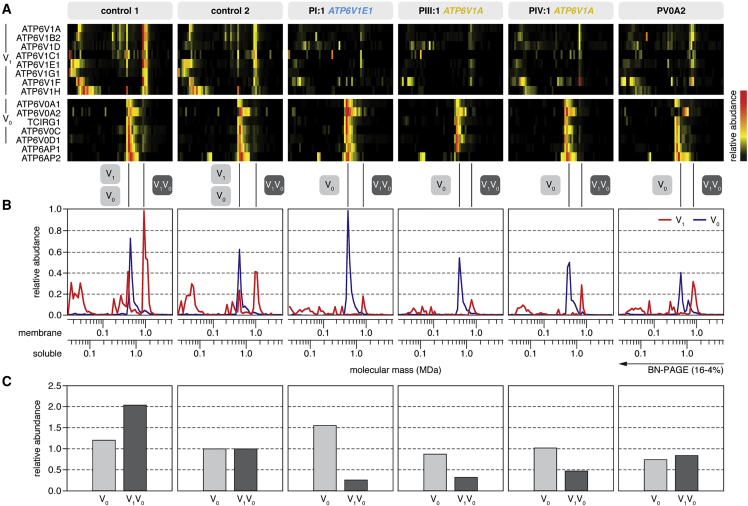
Figure 3.
Complexome Profiling
(A) Heatmap representations of the migration profiles of the identified V1 and V0 subunits and two V-ATPase assembly factors (ATP6AP1 and ATP6AP2) were created by hierarchical clustering and, for proteins that were not grouped together by the clustering algorithm, by correlation profiling. In control fibroblast cultures, the fully assembled V-ATPase and the separate V1 and V0 domains migrated with apparent molecular masses of 1,000, 600, and 450 kDa, respectively. The majority of V1 subunits were integrated in the complete V-ATPase complex, whereas the vast majority of V0 subunits were integrated in the V0 subassembly. In fibroblast cultures from individuals with ATP6V1E1 or ATP6V1A mutations, the abundance of V1 subunits was markedly lower than in control individuals.
(B) Migration profiles of the V1 (red) and V0 (blue) domains show the average value of the abundance of the detected subunits of the respective fraction plotted against the molecular mass.
(C) The amount of fully assembled V-ATPase complex was markedly reduced in fibroblast cultures from all individuals with ATP6V1E1 or ATP6V1A mutations, but the amount of V0 domain was unchanged. In PV0A2, an individual with compound-heterozygous ATP6V0A2 mutations, the amount of fully assembled V-ATPase complex was only moderately reduced. To calculate the values, we summarized the abundances of all detected subunits in the respective fraction and normalized them to control 2.