Fig. 6.
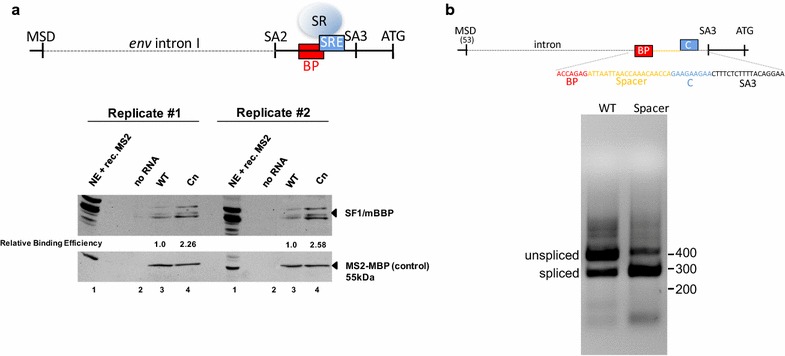
Analyses of wild-type and Cn SA3 recognition by determination of RNA-bound spliceosomal SF1/mBBP. a In vitro synthesized and immobilized RNAs encoding a M2 stem loop followed by wild-type or Cn sequences were incubated with HeLa nuclear extracts. Recombinant MS2-MBP (1 µg) was added to monitor immobilized RNA amounts. Bound proteins were visualized by Western blotting analyses using SF1/mBBP or MBP-antibodies and quantified. Lane 1 represents 10% of the load. b Separation of BP and SRE sequences by a spacer enhances SA3 splicing. LTR and SA3 were cloned in pGL3. A spacer of 20 nucleotides separated BP and the C element and splicing was analysed by RTPCR. A scheme of the spacer plasmid is depicted above the panel. SR serine-arginine-rich, protein, SRE splice regulatory sequence
