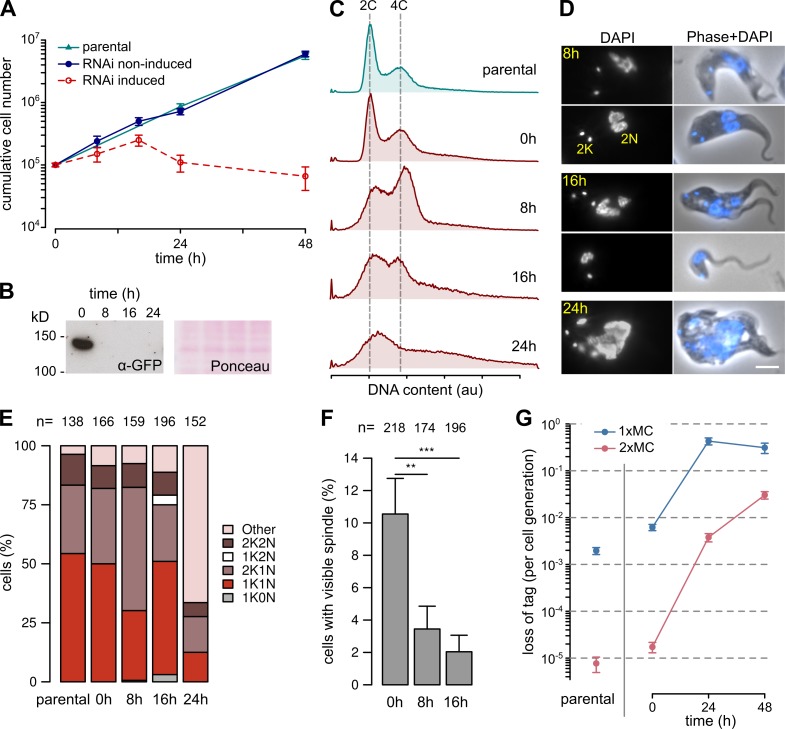
Figure 3.
KKIP1 is essential for chromosome segregation and spindle function. (A) Cell growth in cultures after RNAi-induced ablation of KKIP1. Error bars show SEM (n = 3; P < 0.001 for induced versus noninduced cell numbers at points after 20 h; Student's t test). (B) Immunoblots of cells expressing YFP-KKIP1 showing depletion of protein. Protein loading is shown by Ponceau S stain. (C) Flow cytometry showing disruption of DNA content caused by KKIP1 depletion (representative data from two repeats shown). (D) Phenotypic changes to cells upon RNAi. Bar, 4 µm. (E) Morphological analysis of cell cycle on RNAi shows a buildup of undivided cells with late morphology (2K1N), then formation of cells with aberrant numbers of kinetoplasts (K) and nuclei (N). (F) Loss of visible spindles on KKIP1 depletion, judged by immunofluorescence against β-tubulin (**, P < 0.01; ***, P < 0.001; Student's t test). (G) RNAi-stimulated loss of one or two minichromosomes (MC) from cells. Error bars represent standard error estimates from counts of resistant cells (n = 15–44; P < 0.001 for 24 and 48 h vs. 0 h; Z-test). au, arbitrary units.