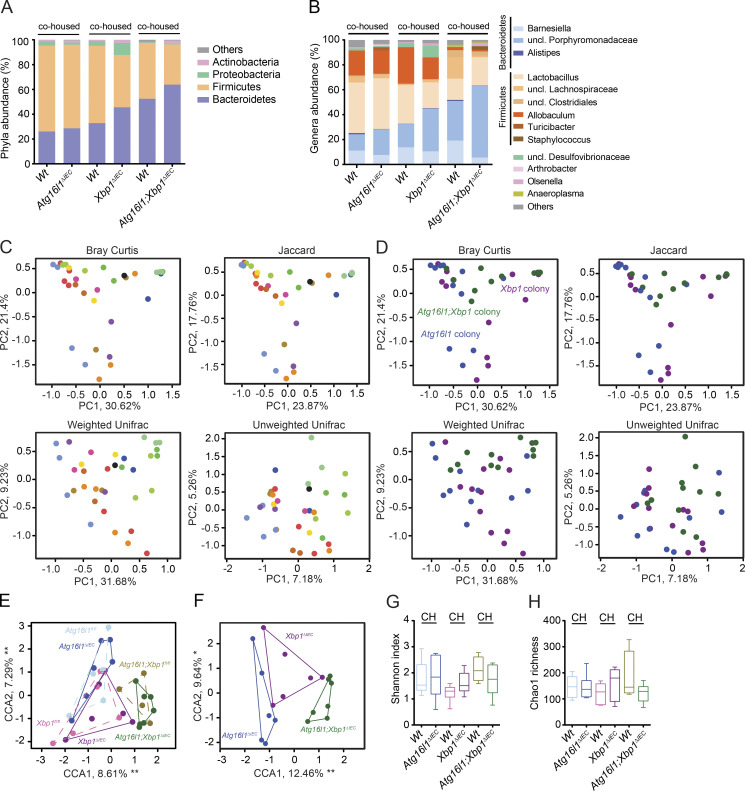
Figure 7.
Atg16l1;Xbp1ΔIEC mice harbor a dysbiotic and transmissible microbiota. (A and B) Relative abundance of microbial taxa at phylum level (≥1%; A) and genus level (≥1%; B) representing ileal mucosa–adherent microbiota of 18-wk-old cohoused mutant (V-cre+) and Wt littermates (V-cre–). n = 5/6/6/6/4/7. uncl., unclassified. (C and D) Ordination analysis of the constraining factors of the cage effect (each dot represents one mouse; cages are indicated as different colors; C) and genotype effect (each dot represents one mouse; colonies are indicated as different colors; D) in the microbiota analysis of cohoused mice based on principal coordinates analysis (Bray Curtis and Jaccard dissimilarity) and Unifrac (weighted and unweighted). Atg16l1 colony, n = 11; Xbp1 colony, n = 12; Atg16l1;Xbp1 colony, n = 11. (E) CCA plot showing a significant genotype effect (F = 2.2270; P = 0.001) and gender effect (F = 2.2639; P = 0.004) in the indicated groups (n = 5/6/6/6/4/7). Significant genotype effect is shown in D. Cage and litter effects were partialled out in this analysis and are shown in C. (F) CCA plot of knockouts only (n = 6/6/7) constrained for mutation and gender effect (cage and litter effect were partialled out). (G and H) α diversity estimates Shannon’s index analyzing species evenness (G), and Chao 1 determining species richness (H) is shown (n = 5/6/6/6/4/7). Statistical analysis was performed using nonparametric Kolmogorov-Smirnov test. CH, cohoused.