Fig. 8.
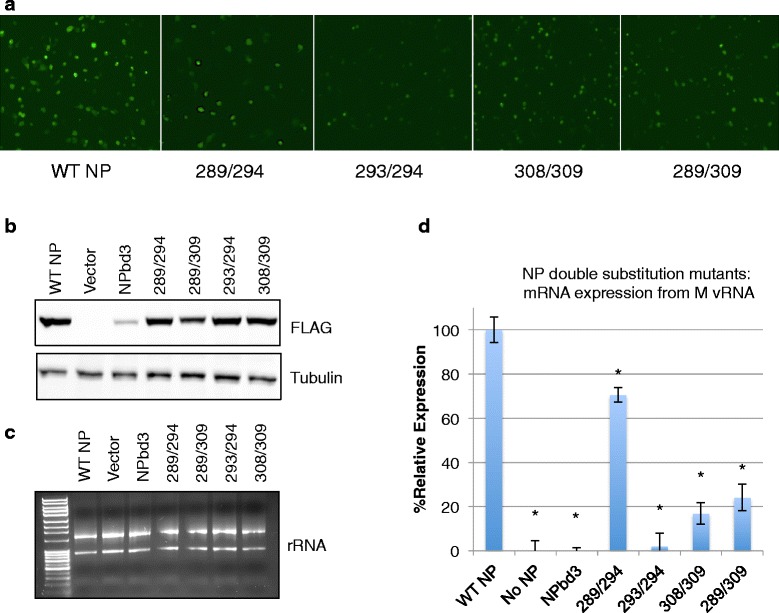
NP double amino acid substitution mutants display decreased activity in reconstituted vRNP assay. a Cells expressing reconstituted vRNPs with GFP-M vRNA were visualized for GFP expression with a Nikon Eclipse TS100 (Nikon Intensilight C-HGFI for fluorescence) inverted microscope and images captured with the Nikon DS-Qi1Mc camera with NS Elements software. Number indicates the two NP residues substituted with glycine. b Total protein was isolated from cells expressing reconstituted vRNPs with GFP-M vRNA template and the indicated NP mutant. Total protein extract was separated on 10% SDS PAGE and transferred to nitrocellulose. WT NP, NPbd3, and NP double mutants were detected with anti-FLAG. Anti-tubulin was used as loading control. Expected size was confirmed by comparison with Fisher BioReagents EZ-Run protein standards. c RNA from cells expressing reconstituted vRNPs with FLAG-M vRNA and either WT NP, no NP, or NPbd3, or the double amino acid mutants were isolated and analyzed by 1% bleach/agarose gel electrophoresis. d One microgram of RNA was treated with DNase before being reverse transcribed with oligo dT (mRNA) and analyzed through quantitative PCR with primers targeting the M gene. qPCR reactions were carried out in triplicate. Significance was evaluated through t-test by comparing WT NP; asterisks indicate p-values <0.02
