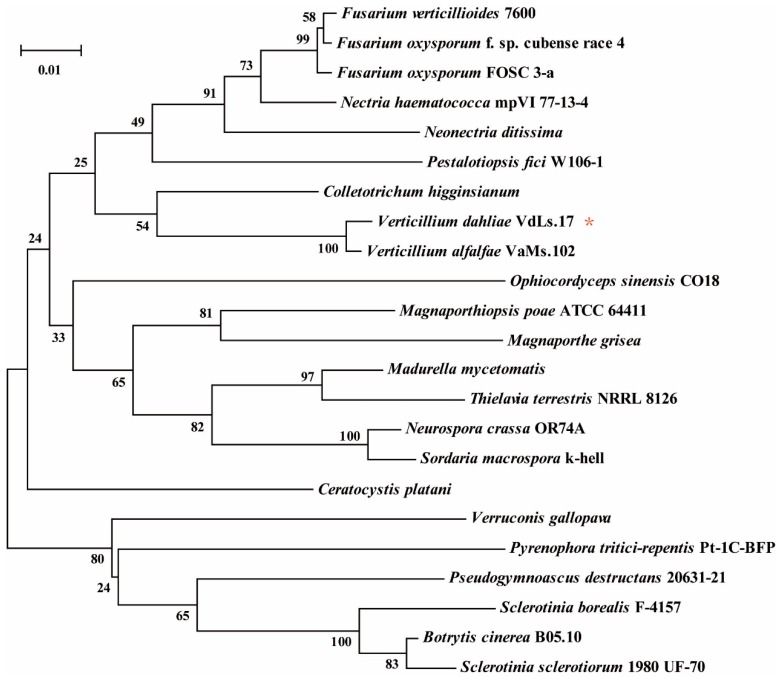
Figure 1.
Phylogenetic analysis of AAC amino acid sequences from different fungal species. The phylogenetic tree was constructed by MEGA software (version 6.06; bootstraps: 1000). Fungal species and protein accession numbers: Verticillium dahliae VdLs.17 (XP_009654735.1); Nectria haematococca mpVI 77-13-4 (XP_003051617.1); Fusarium oxysporum FOSC 3-a (EWZ02370.1); Verticillium alfalfae VaMs.102 (XP_003004480.1); Fusarium verticillioides 7600 (EWG42987.1); Pestalotiopsis fici W106-1 (XP_007835574.1); Fusarium oxysporum f. sp. cubense race 4 (EMT68221.1); Magnaporthiopsis poae ATCC 64411 (KLU91586.1); Neonectria ditissima (KPM44251.1); Madurella mycetomatis (KOP45712.1); Colletotrichum higginsianum (CCF32866.1); Ceratocystis platani (KKF94862.1); Pseudogymnoascus destructans 20631-21 (XP_012739498.1); Botrytis cinerea B05.10 (XP_001559435.1); Sclerotinia sclerotiorum 1980 UF-70 (XP_001598713.1); Verruconis gallopava (KIW06207.1); Sclerotinia borealis F-4157 (ESZ96107.1); Neurospora crassa OR74A (XP_011393638.1); Sordaria macrospora k-hell (XP_003351160.1); Pyrenophora tritici-repentis Pt-1C-BFP (XP_001934086.1); Ophiocordyceps sinensis CO18 (EQK99145.1); Magnaporthe grisea (AAX07662.1); Thielavia terrestris NRRL 8126 (XP_003658188.1). * represents the query sequence.